
METTL7B (methyltransferase-like 7B) identification as a novel biomarker for lung adenocarcinoma
Introduction
Lung cancer is a significant cause of cancer-related mortalities over the world (1). Statistical data show annual 2.1 million new lung cancer cases and 1.8 million deaths worldwide, corresponding to approximately 1 in 5 (18.4%) cancer deaths (2). The most common histological subtype of lung cancers is lung adenocarcinoma (LUAD), consisting of 40% of all lung cancers (3-7). Even though survival rate is improving in 60% of LUAD patients with targetable gene alterations, still the mortality is high because of a delayed identification time, the progress of cancer to advanced stages with widespread metastasis, and poor drug outcomes (1,8,9). The overall 5-year survival rate for LUAD is less than 20% owing to untimely diagnosis (10-13). Regardless of many improvements and accomplishments in the basic research concerning LUAD, the underlying molecular mechanisms have not been fully elaborated. The lack of specific and sensitive biomarkers that can prove significant in its early diagnosis still is undiscovered. Therefore, it is essential to find novel biomarkers that can detect cancer in its initial stages.
Methyltransferase-like 7B (METTL7B) is also known as Associated with Lipid Droplet Protein 1 (ALDI). Its gene is found on chromosome 12 with a molecular mass of 27.775 kDa. This protein has not been studied thoroughly; therefore, its function remains unknown. Studies have suggested a significant role of METTL7B in tumorigenesis in breast and thyroid cancer (14,15). Ye and colleagues have reported, the upregulation of METTL7B in papillary thyroid cancer (PTC), and its role in promoting invasion and migration of cancer via epithelial-mesenchymal transition (EMT) (16).
METTL7B correlation with some other diseases have been reported, such as severe preeclampsia (17), non-alcoholic steatohepatitis lipid metabolism (18), and infections (19). However, whether METTL7B plays a role in lung cancer has been studied rarely. In the current study, we found that METTL7B was upregulated in LUAD from two dependent cohorts, in surgically resected tissues, and serum samples, suggesting that METTL7B could be a potential biomarker for LUAD. Moreover, further exploration showed that METTL7B has a significant effect on cell proliferation, migration, and invasion of LUAD.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-4574).
Methods
Human tissue microarray
Tissue microarrays of LUAD (HLugA180Su07, Outdo Biotech, Shanghai, China) consisted of surgically resected 96 LUAD tissue and 77 matched adjacent non-diseases tissue specimens. The median age was 60.1 years, the range was 25–84 years, and 40 were females from the two tissue array blocks and were retrieved between September 2004 and June 2009. 7th edition International Union Against Cancer/American Joint Committee on Cancer TNM classification was used to diagnose and stage all the cases. Those patients were followed-up between 36- and 96-month post-surgery.
Immunohistochemistry (IHC)
IHC was done to examine METTL7B’s expression in tissue samples. The tissue sections were deparaffinized, rehydrated, and then incubated with 3% hydrogen peroxide (H2O2) in methanol. Antigen retrieval was conducted by using ethylene-diamine-tetraacetic acid (EDTA) buffer. The tissue sections were blocked with 5% bovine serum (BSA) and then probed with anti-METTL7B (1:50, Protientech Wuhan, China, #17001-1-AP) at 4 °C overnight. Biotinylated secondary antibodies were added to the tissue sections, detected by the Streptavidin-Peroxidase IHC assay kit and diaminobenzidine (DAB) (ZSGB-bio, Beijing, China). Two independent pathologists evaluated the immunostaining in a blinded fashion and scored, corresponding to staining intensity and the percentage of stained cells. Staining intensity score was given as negative staining: 0 point; weak staining: 1 point; moderate staining: 2 points; and strong staining: 3 points. Percentage of stained cells was quantified as cells 1: ≤25%; 2: 26–50%; 3: 51–75%; and 4: ≥75%. The immunoreactivity score was calculated by multiplying the staining intensity score and the percentage of positive cells. According to the immunoreactivity score of METTL7B, the sample was graded into high expression (a score of ≥4) or low expression (<4). The positive control of METTL7B expression was set up, referring to lung cancer from the website (Http://www.proteinatlas.org).
Serum samples
Serum samples were collected from 37 females and 28 males, the median age was 57 years and the age range was 24–83 years. Serum samples were harvested following a standard operating procedure. In brief, peripheral blood (2 mL each subject) was collected in a serum separator tube (SST) and allowed to clot for 30 minutes before centrifugation for 15 minutes at 1,000 g/minutes. Immediately after centrifugation, serum was transferred to clean polypropylene tubes and stored at −80 °C. Written informed consent was retrieved from all participants. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Review Committee of the Second Hospital of Dalian Medical University. Reference number for approval was 2019046.
Enzyme-linked immunosorbent assay (ELISA)
Sandwich ELISA method was used to measure the METTL7B levels in the cell culture supernatant and clinical serum samples using the kits from Omnimabs (Omnimabs, California, United States of America (USA). ELISA Assay kit used purified antibody immobilized onto the surface of the microtiter well plates. The samples, including standards, were added into antibody-coated microtiter wells via the pipette. Following the incubation and washing of the wells, biotinylated antibody and combined Streptavidin- horseradish peroxidase (HRP) were added and again incubated. A complex is formed between Antibody-Antigen-Enzyme linked Antibody. After another washing step, the tetramethylbenzidine (TMB) substrate solution is added to the well plates. Ammonium sulfate (NH4)2SO4 was used to stop the reaction and colorimetric change observed. The color change was read at 450 nm on a microplate reader. The optical density (OD) values were directly proportional to Protein concentration. A standard curve was used to calculate protein concentrations.
Cell culture
Human LUAD cell lines H1299, H1975, A549, H827, PC9 and SPCA1, and the normal human lung epithelial cell BEAS2B, were retrieved from the Chinese Academy of Medical Sciences (Beijing, China). All cell lines were routinely cultured in the Roswell Park Memorial Institute (RPMI 1640) medium, holding 10% fetal bovine serum (FBS) at 37 °C, holding 5% carbon dioxide (CO2).
Quantitative-polymerase chain reaction (q-PCR)
Total Ribonucleic acid (RNA) was extracted using the Trizol® reagent (Transgen biotech, Beijing, China) and quantitated at OD260 nm. Total RNA (1.0 µg) was treated with RNase-free DNase I and reverse-transcribed into complementary deoxyribonucleic acid (cDNA) using random primers and Superscript II® retrotranscriptase (Invitrogen, California, USA). The resulting cDNAs were mixed with the synergy brands, Inc (SYBR) polymerase chain reaction (PCR) master mix (Applied Biosystems, Waltham, USA) and run on the StepOnePlus Applied Biosystems Realtime PCR machine. One cycle of a denaturing step (3 minutes at 95 °C) was applied, which was followed by 40 cycles of amplification (12 s at 95 °C, the 30 s at 62 °C and 30 s at 72 °C), with fluorescence measured during the extension. Primers used in this study are as follows:
GAPDH: 5'-CATGAGAAGTATGACAACAGCCT-3' (forward); 5'-AGTCCTTCCACGATACCAAAGT-3' (reverse); METTL7B: 5'-CCAGATAAAGGGGCTTACAGGAG-3' (forward); 5'-TCAGCCATGCTCTTTGTCAGG-3' (reverse).
Western blot
Western blot procedure was performed, supporting the standard protocol. Cells were lysed with radioimmunoprecipitation assay (RIPA) buffer (Sigma, Saint Louis, USA) along with protease inhibitor phenylmethylsulfonyl fluoride (PMSF) (Sigma, Saint Louis, USA) and centrifuged to extract the protein. We used the bicinchoninic acid assay (BCA) protein kit (Thermo Fisher Scientific Inc., Waltham, USA) to measure the concentration of protein. Extracted protein was subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Millipore, Billerica, USA). To block the membrane, a 5% defatted milk solution in 0.05% Tris-buffered saline/Tween-20 (TBST) was used and then incubated with anti-METTL7B rabbit polyclonal antibody (1:500; Proteintech, Wuhan, China, #17001-1-AP) at 4 °C overnight. The next morning membrane was washed with 0.05% TBST and incubated with secondary antibody conjugated with horseradish peroxidase (1:5,000; Proteintech, Wuhan, China). The enhanced chemiluminescence (ECL) western blotting substrate was used to visualize the chemiluminescence of the blots (NCM Biotech, Suzhou, China). Image J software (National Institutes of Health, USA) was used to detect Protein expression. An anti-GAPDH (Glyceraldehyde-3-phosphate dehydrogenase) rabbit monoclonal antibody (1:2,000, Proteintech, USA) was used as the loading control. The tests were repeated thrice to decrease the variability.
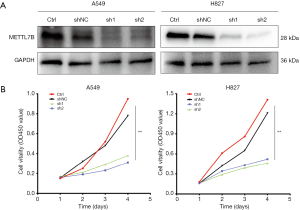
Short hairpin ribonucleic acid (shRNA)transfection
The LV17 lentiviral vector (GenePharma, Suzhou, China) was used to construct METTL7B-targeting short hairpin ribonucleic acid (shRNA), and lentiviral particles expressing luciferase-enzyme infused shRNA were packaged in 293T cells. The sequences of shRNAs were 5'-GCGGAACCGGAGCCAACUUTT-3' (sh1) and 5'-GCAGGAGGUCCGGAGAGUATT-3' (sh2). Luciferase infused shRNA, which does not target any gene product, was used as a short hairpin negative control (shNC). A549 and H827 cells were transduced with lentivirus shRNA1, shRNA2, and shNC and treated with polybrene to precipitate stable knockdown of METTL7B. The Western blot method measured the silencing effect of the METTL7B gene.
Cell proliferation assay
The cell proliferation was detected by cell counting kit-8 (CCK-8) assay kit. Following the standard protocol, the transfected tumor cells (H827 3×103 cells/well; A549 2×103 cells/well) and negative control (NC) cells were seeded into a flat bottomed 96-well plate. The plate was divided into four sections and incubated at 37 °C with 5% CO2. CCK-8 reagent (APExBIO, Houston, Texas, USA) was added to the wells of each section and then incubated for 2 hours. The absorbance was recorded at 450 nm. This process was repeated at these time-points 0, 24, 48, and 72 hours.
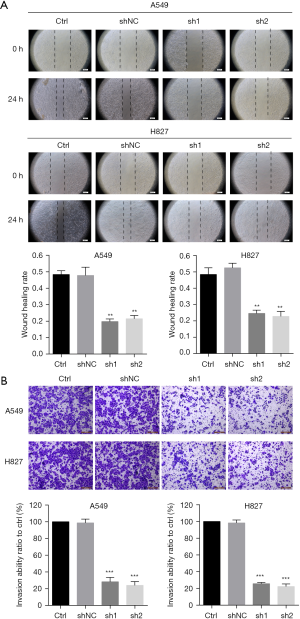
Wound closure assay
For the Wound closure assay, cells were cultured in 6-well plates. After the cells reached a 90% confluence, cells were subjected to serum starvation for all night. Cells were scratched with a 200 µL pipette tip and then cultured in a 0.5% serum-containing medium. Images were recorded at 0 and 24 hours post wound initiation with the application of an inverted microscope.
Invasion assay
The cell suspension holding (2.5×105 cells/well) in serum-free media was seeded onto the Matrigel-coated Transwell chambers for the invasion assay. The Transwell chambers were placed into the 24-well plate (8-µm pore, Corning), holding 20% FBS medium and cultured for 24 hours. A cotton swab was used to remove the non-invading cells present on the superior surface of the top chamber. The invading cells present on the bottom surface of the top chamber were fixed with 4% paraformaldehyde, stained with 2% crystal violet, imaged under a microscope (Leica, TCSSP5II, Wetzlar, Germany). The randomly selected invading cells were counted in five fields per insert in a blinded manner.
Statistical analysis
The statistical analysis was performed using GraphPad Prism (version 8.0; GraphPad Software, San Diego, CA, USA). The Data was expressed as mean ± standard deviation. Student’s t-test, analysis of variance (ANOVA), or Chi-square test were applied to compare the data among different experimental groups. The survival curve was formulated and matched with the log-rank test using the Kaplan-Meier method. A P value of less than 0.05 was considered statistically significant.
Results
Overexpression of METTL7B in clinical LUAD and associated with poor prognosis
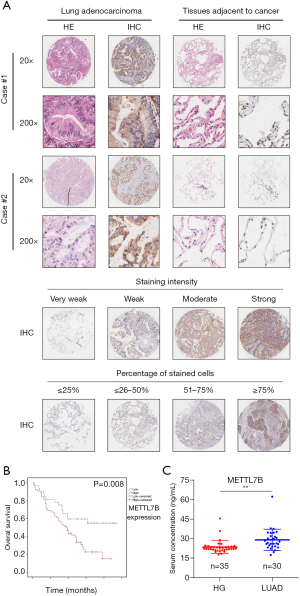
Tissue microarray (TMA) holding LUAD tumor samples (n=96) and matched adjacent non-tumor tissues (n=77) were utilized for IHC staining to determine METTL7B’s expression in clinical LUAD patients. IHC results showed that METTL7B staining was notably dominant in the cytoplasm of LUAD tissue, and METTL7B’s expression was significantly higher in LUAD tissues than in the matched non-tumor tissues (Figure 1A). Further quantitative analysis showed that almost 70.13% (54/77) of LUAD tissues expressed elevated levels of METTL7B compared with 7.79% (6/77) of adjacent non-tumor tissues (P<0.001) (Table 1).

Full table
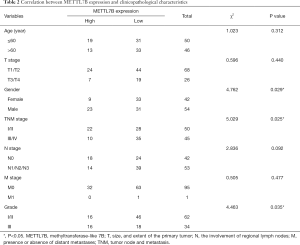
The clinicopathologic characteristics and prognostic information of TMA patients were collected to explore the association between METTL7B and outcomes of LUAD patients. Stratification analysis showed that higher METTL7B expression in LUAD was significantly correlated with sex (P=0.029), TNM stage (P=0.025), and Grade (P=0.035) (Table 2). Kaplan-Meier survival analysis showed that LUAD patients with higher expression of METTL7B had a significantly reduced survival rate (P<0.05) (Figure 1B).
Full table
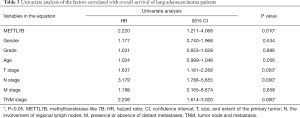
In the univariate analysis of the factors correlated with the overall survival of LUAD patients, high METTL7B expression was significantly associated with poor overall survival (HR =2.220, 95% CI: 1.211–4.086; P=0.010), along with the advanced TNM stages (HR =2.208, 95% CI: 1.614–3.020; P=0.000), positive lymph node metastasis (HR =3.179, 95% CI: 1.788–5.655; P=0.000) and tumor size (HR =1.637, 95% CI: 1.181–2.268; P=0.003) (Table 3). Therefore, the results suggest, METTL7B overexpression was associated with poor prognosis of patients in LUAD.
Full table
Since METTL7B could be secreted to the extracellular region, we further determined the diagnostic value of serum METTL7B levels for LUAD. We collected serum samples from the other dependent cohort, which had the healthy control group (n=35) and the LUAD group (n=30). ELISA results showed that the average METTL7B level of the LUAD group was significantly higher than the healthy control group, confirming the diagnostic value of serum METTL7B levels in LUAD (Figure 1C). In conclusion, our findings confirm that METTL7B is overexpressed in clinical serum and tumor tissues of LUAD patients, being identified as a diagnostic serum marker for LUAD, and it is associated with poor prognosis.
Validation of METTL7B overexpression in LUAD cell lines
To further confirm our findings of METTL7B overexpression, we performed qPCR and western blot to evaluate the METTL7B level at mRNA and protein level in cell lines. Six human LUAD cell lines (H1299, H1975, A549, H827, PC9, and SPCA1) and the normal human lung epithelial cell (BEAS2B), which acted as the control group were involved in the experiment. The results of qPCR and western blot assays were in accordance, revealing a significantly higher METTL7B level in all LUAD cell lines in comparison with the normal human lung epithelial cell (Figure 2A,B)
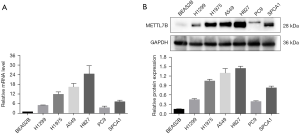
The western blot revealed the upregulation of METTL7B in all LUAD cell lines when compared with the normal control lung cell line (Figure 2B).
A549 and H827 cell lines showed high endogenous METTL7B expression in this group and, therefore, were selected for further study. In short, overexpression of METTL7B was often found in LUAD.
Inhibition of METTL7B suppressed LUAD cell proliferation
The METTL7B gene was silenced by using lentivirus-mediated two different short hairpin RNAs (shRNA1 and shRNA2) in H827 and A549 cells, which showed high endogenous METTL7B expression to ratify further whether METTL7B gene silencing could inhibit cell proliferation. Western blotting and CCK-8 were performed respectively to evaluate the efficiency of knockdown and assess cell proliferation. Both shRNAs effectively knocked down METTL7B resulting in decreased expression in the LUAD cell lines, as shown in western blot (Figure 3A). With METTL7B’s depletion, cell proliferation was significantly decreased as compared to the control (Figure 3B). These findings show that METTL7B overexpression augmented cell proliferation in LUAD.
METTL7B promotes LUAD cell migration and invasion
To evaluate the effect of METTL7B on LUAD cell migration and invasion, we performed wound healing assay and invasion assay, respectively. Our results showed that METTL7B knockdown led to profound impairment in cell migration ability of A549 and H827 relative to the control (Figure 4A). In vitro invasion, assays showed that METTL7B depletion resulted in conspicuous suppression of LUAD cell invasion capability (Figure 4B). Hence, our results showed the significance of METTL7B in LUAD cell migration and invasion.
Discussion
LUAD is considered to be the most frequent pathological subtype of Lung cancer in females and non-smokers (20-22). LUAD progression is a multistep process consisting of a premalignant lesion, carcinoma in situ followed by the invasive phenotype of LUAD (23). Owing to the high morbidity and mortality of LUAD, it is urgent to find new diagnostic methods and clinical management.
METTL7B belongs to the methyltransferase-like proteins (METTL) family. METTL family consists of over twenty-seven members, but their functions have not yet been explored thoroughly (24-27). METTL7A is a closely related protein with METTL7B sharing 59% sequence identity and more widely studied than METTL7B. It is proved to play an essential role in hepatocellular carcinoma (HCC), thyroid cancer, LUAD, and breast cancer (14,28-30). Recently METTL7B has received much more attention. Originally, METTL7B was discovered as a Golgi-interrelated methyltransferase with unidentified functions (31). Initially, it was reported to be localized to the Golgi complex, but later on, it was detected localized to endoplasmic reticulum (ER) in Hela cells and lipid bodies in the liver (32). To our knowledge, METTL7B was associated with diseases such as severe preeclampsia, non-alcoholic steatohepatitis lipid metabolism, and infections (17-19). Increasing evidence defined the role of METTL7B in the progression of the malignant tumors including primary thyroid cancer (PTC), breast tumor and non-small cell lung cancer (NSCLC). In PTC, METTL7B was upregulated and facilitated thyroid cancer cell migration and invasion via promoting tumor growth factor beta-1 (TGF- β1) induced EMT (16). In breast cancer, RhoBTB (BTB stands for broad-complex, tramtrack, bric à brac) silencing led to the fragmentation of the Golgi complex because of decreased METTL7B expression, which enhanced the invasion of breast cancer cells (14). In lung cancer, a recent study demonstrated that METTL7B was overexpressed in NSCLC tumor tissues and promoted tumorigenesis by regulating cell cycle progression (33). However, the diagnostic value of METTL7B as a potential biomarker for lung cancer required further confirmation and the role of METTL7B in invasion and metastasis of lung cancer had not been explored.
In our study, we determined METTL7B’s function as a potential oncogene in tumor development and LUAD’s progression. We showed that METTL7B was frequently upregulated in LUAD tissues compared to the matched normal control tissue samples with IHC staining and was associated with a reduced overall survival rate in LUAD patients. Also, IHC staining on tissue microarrays was significantly correlated with tumor size, advanced TNM stages, and lymph node metastasis, suggesting that overexpression of METTL7B in LUAD may promote the metastatic phenotype. Further, in vitro assays showed that METTL7B gene silencing produced apparent hindrance in malignant properties of LUAD cells, such as inhibition of tumor cell proliferation, invasion, and migration. Therefore, it is noteworthy that these findings suggested that METTL7B had significant involvement in LUAD progression. Besides, we also supplied the experimental evidence of METTL7B to serve as a potential serum diagnostic biomarker since METTL7B could be secreted in the extracellular region. The elevated levels of METTL7B in the clinical serum of LUAD patients were compared to healthy volunteers, providing an essential gateway to non-surgical detection methods rendering ease in clinical diagnosis. The findings that METTL7B may serve as a potential novel biomarker for the diagnosis and prognosis of LUAD as well as the important role of METTL7B in migration and invasion of LUAD are unique to our study.
In short, METTL7B could act as a serum biomarker for LUAD diagnosis and a potentially useful therapeutic target in LUAD therapy since it was defined that METTL7B took part in the tumor progression.
Acknowledgments
Funding: This study was funded in part by the National Natural Science Foundation of China (No. 81972916, 81502702).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-4574
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-4574
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4574). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Written informed consent was retrieved from all participants. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Review Committee of the Second Hospital of Dalian Medical University. Reference number for approval was 2019046.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Shiraishi K, Kunitoh H, Daigo Y, et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet 2012;44:900-3. [Crossref] [PubMed]
- Mao H. Clinical relevance of mutant-allele tumor heterogeneity and lung adenocarcinoma. Ann Transl Med 2019;7:432. [Crossref] [PubMed]
- Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Walters S, Maringe C, Coleman MP, et al. Lung cancer survival and stage at diagnosis in Australia, Canada, Denmark, Norway, Sweden and the UK: a population-based study, 2004-2007. Thorax 2013;68:551-64. [Crossref] [PubMed]
- Maemura K, Watanabe K, Ando T, et al. Altered editing level of microRNAs is a potential biomarker in lung adenocarcinoma. Cancer Sci 2018;109:3326-35. [Crossref] [PubMed]
- Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998-2006. [Crossref] [PubMed]
- Buss EJ, Wang TJC. Treatment of lung adenocarcinoma brain metastases: what is the role of radiotherapy in the age of precision medicine? Transl Lung Cancer Res. 2018;7:S318-20. [Crossref] [PubMed]
- Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012;150:1107-20. [Crossref] [PubMed]
- Yang M, She Y, Deng J, et al. CT-based radiomics signature for the stratification of N2 disease risk in clinical stage I lung adenocarcinoma. Transl Lung Cancer Res. 2019;8:876-85. [Crossref] [PubMed]
- Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014;14:535-46. [Crossref] [PubMed]
- Xu SB, Xie MR, Li CW, et al. Correlation of pulmonary venous circulating tumor cells with clinicopathological parameters in patients with early-stage lung adenocarcinoma. Transl Cancer Res 2019;8:887-98. [Crossref]
- McKinnon CM, Mellor H. The tumor suppressor RhoBTB1 controls Golgi integrity and breast cancer cell invasion through METTL7B. BMC Cancer 2017;17:145. [Crossref] [PubMed]
- Cai WY, Chen X, Chen LP, et al. Role of differentially expressed genes and long non-coding RNAs in papillary thyroid carcinoma diagnosis, progression, and prognosis. J Cell Biochem 2018;119:8249-59. [Crossref] [PubMed]
- Ye D, Jiang Y, Sun Y, et al. METTL7B promotes migration and invasion in thyroid cancer through epithelial-mesenchymal transition. J Mol Endocrinol 2019;63:51-61. [Crossref] [PubMed]
- Nevalainen J, Skarp S, Savolainen ER, et al. Intrauterine growth restriction and placental gene expression in severe preeclampsia, comparing early-onset and late-onset forms. J Perinat Med 2017;45:869-77. [Crossref] [PubMed]
- Thomas A, Klein MS, Stevens AP, et al. Changes in the hepatic mitochondrial and membrane proteome in mice fed a non-alcoholic steatohepatitis inducing diet. J Proteomics 2013;80:107-22. [Crossref] [PubMed]
- Abdel-Hameed EA, Ji H, Sherman KE, et al. Epigenetic modification of FOXP3 in patients with chronic HIV infection. J Acquir Immune Defic Syndr 2014;65:19-26. [Crossref] [PubMed]
- Torres-Duran M, Barros-Dios JM, Fernandez-Villar A, et al. Residential radon and lung cancer in never smokers. A systematic review. Cancer Lett 2014;345:21-6. [Crossref] [PubMed]
- Hsu CL, Chen KY, Shih JY, et al. Advanced non-small cell lung cancer in patients aged 45 years or younger: outcomes and prognostic factors. BMC Cancer 2012;12:241. [Crossref] [PubMed]
- Couraud S, Zalcman G, Milleron B, et al. Lung cancer in never smokers--a review. Eur J Cancer 2012;48:1299-311. [Crossref] [PubMed]
- Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol 2011;29:2972-7. [Crossref] [PubMed]
- Wang X, Feng J, Xue Y, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 2016;534:575-8. [Crossref] [PubMed]
- Pendleton KE, Chen B, Liu K, et al. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017;169:824-35 e14.
- Lin S, Choe J, Du P, et al. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell 2016;62:335-45. [Crossref] [PubMed]
- Ignatova VV, Jansen P, Baltissen MP, et al. The interactome of a family of potential methyltransferases in HeLa cells. Sci Rep 2019;9:6584. [Crossref] [PubMed]
- Zhou S, Shen Y, Zheng M, et al. DNA methylation of METTL7A gene body regulates its transcriptional level in thyroid cancer. Oncotarget 2017;8:34652-60. [Crossref] [PubMed]
- Qi L, Song Y, Chan THM, et al. An RNA editing/dsRNA binding-independent gene regulatory mechanism of ADARs and its clinical implication in cancer. Nucleic Acids Res 2017;45:10436-51. [Crossref] [PubMed]
- Guo T, Ma H, Zhou Y. Bioinformatics analysis of microarray data to identify the candidate biomarkers of lung adenocarcinoma. PeerJ 2019;7:e7313. [Crossref] [PubMed]
- Wu CC, MacCoss MJ, Mardones G, et al. Organellar proteomics reveals Golgi arginine dimethylation. Mol Biol Cell 2004;15:2907-19. [Crossref] [PubMed]
- Turro S, Ingelmo-Torres M, Estanyol JM, et al. Identification and characterization of associated with lipid droplet protein 1: A novel membrane-associated protein that resides on hepatic lipid droplets. Traffic 2006;7:1254-69. [Crossref] [PubMed]
- Liu D, Li W, Zhong F, Yin J, Zhou W, Li S, et al. METTL7B Is Required for Cancer Cell Proliferation and Tumorigenesis in Non-Small Cell Lung Cancer. Front Pharmacol. 2020;11:178. [Crossref] [PubMed]
(English Language Editor: J. Chapnick)


