
Identification of critical radioresistance genes in esophageal squamous cell carcinoma by whole-exome sequencing
Introduction
Esophageal carcinoma (EC) is the seventh most common and the sixth most lethal cancer worldwide (1). Esophageal squamous cell carcinoma (ESCC) is the most common histology and is endemic in East Asia. The prognosis of EC is abysmal, and the overall 5-year survival rate is approximately 20% (2). Immunotherapy is a promising treatment for ESCC, but the effective rate is still low, and most of it is used in the second line of advanced esophageal cancer. In China, ESCC ranks fourth in cancer-related deaths, with an estimated case number of over 375,000 deaths from EC occurring in 2015 (3).
Radiotherapy (RT) has historically played a pivotal role in the management of EC; however, the efficacy of radiation monotherapy is inferior. Radioresistance results in high locoregional recurrence and metastasis (4), possibly the chief reasons for its poor efficacy (5). For decades, many related studies have been performed (6-9). In the era precision medicine, different from cancer species that could be treated with targeted drugs, due to insufficient of biomarker assays to predict which patients are radiosensitive and which are radioresistant, radioresistance-associated predictive biomarkers of ESCC are more urgently needed.
As a complex disease with multiple genes, the pathogenesis of tumor may be related to the variation of exon group. Whole-exome sequencing (WES) technology has the advantages of low cost and high efficiency, which makes it have considerable application prospect in the study of disease mechanism (10). After introducing of WES, many driver genes of ESCC pathogenesis and several important signaling pathways have been identified (11-15). However, radioresistance is a complex phenomenon involving cancer stem cells (CSCs), intrinsic radiosensitivity, human papillomavirus (HPV) status, hypoxia, and repopulation (16). Subsequent studies attempting to identify radioresistance-associated genes in the treatment-naïve setting are unlikely to be successful because various factors are expected to affect the radiation response. However, for postoperative patients treated with esophageal resection and en bloc lymphadenectomy, only a few remaining tumor cells and other confounding factors are expected. Comparing the sequencing data of postoperative ESCC patients with opposite prognoses who received postoperative radiotherapy (PORT) is a more suitable approach, and the effects of genetic alterations on radioresistance can be more easily distinguished.
In this study, two cohorts of ESCC patients who received PORT and had opposite prognoses were set up. We characterized and compared the somatic alterations by WES. This study revealed different mutational signatures and frequently altered genes underlying ESCC radioresistance, easing a comprehensive understanding of radiation oncology.
We present the following article/case in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5196).
Methods
Samples and clinical data
Two independent cohorts, including 23 ESCC patients from the Affiliated Hospital of Nantong University (Jiangsu, China), were enrolled in this study. Cohort 1, the relapsed ESCC group, included 12 patients with an early relapse of ESCC who had received PORT. Cohort 2, the stable ESCC group, included 11 ESCC patients who had received PORT and were confirmed to be in stable condition. Paraffin-embedded tumor samples, adjacent nontumor specimens, and tissues from recurrent tumors of those individuals were obtained from the pathology department. The tumor samples of all affected individuals had a tumor cell content of at least 40–50%. In this retrospective WES study, the inclusion criteria were as follows: (I) all patients had undergone radical surgical treatment and PORT; (II) the histological types of all ESCC cases had been determined; (III) postoperatively, all patients had pathological stage I to III disease according to the American Joint Committee on Cancer (AJCC) 8th edition cancer staging manual; (IV) no patient had proceeded with preoperative neoadjuvant therapy; (V) all patients received an adequate dose of radiation, and recurrence occurred within the radiation field, as confirmed by the attending physician; and (VI) in all patients, recurrence was determined within two years after the end of RT by biopsy, PET/CT or CT. Clinical data of the studied subjects were collected from medical records at the Affiliated Hospital of Nantong University. Patient information was collected via telephone interviews with ESCC-affected individuals or, if these individuals were deceased, through their first-degree relatives via a survey questionnaire.
Approval from the Institutional Research Ethics Committee of Affiliated Hospital of Nantong University (Jiangsu, China) (No. 2019-K078) and prior consent from patients was obtained for using the clinical materials for research purposes. The study was performed following the Declaration of Helsinki (as revised in 2013).
WES and variant interpretation
WES was performed by Geneseeq Inc. (Nanjing, China) on DNA extracted from the qualified genomic DNA from 23 tumors and matched normal tissues. In brief, Illumina paired-end libraries were constructed from DNA samples and subjected to whole-exome capture using the SureSelect Human All Exon V6 + UTR r2 core design [91 megabyte (Mb), Agilent] followed by sequencing on an Illumina HiSeq 4000 platform. The captured, purified, and amplified libraries targeting the exomes from the patients were subjected to paired-end sequencing (2×100 bp) on the Illumina HiSeq 4000 and were aligned to human genome 19 (hg19) using Burrows-Wheeler Aligner (BWA) with default settings. Single-nucleotide variants (SNVs) were defined by the DiBayes algorithm as differences from the reference human genome sequence, and variants present in the paired normal DNA or representing sequencing errors were removed. The SOLiD Small InDel Tool was used to screen small insertions and deletions. We performed ADTEx (17) to detect the somatic copy number variations (SCNVs). ADTEx is tailored for WES data; to infer SCNVs in ESCC genomes from the normalized ratio of WES data from tumor and matched non-neoplastic mucosae. Copy number gains and losses were referred to as amplifications and deletions, respectively.
Statistical analysis
Statistical analyses were performed using R (www.r-project.org) and GraphPad Prism 7.0 (GraphPad Software, Inc., San Diego, CA, USA). Progression-free survival (PFS) was determined using the Kaplan-Meier method, and the log-rank test was used to determine the differences between patients with or without mutations in the specified gene. A Cox proportional hazard regression model was used to analyze the prognostic relevance of the clinical parameters. P<0.05 was considered to indicate a statistically significant difference.
Results
Clinical characteristics of the ESCC patients
In this retrospective study, 23 pairs of tumor-normal PORT ESCC specimens (12 patients with early relapse, 11 individuals in stable condition) and 7 tissues from recurrent tumors were comprehensively characterized via WES. All patients underwent radical resection (R0), subsequent PORT, and selective chemotherapy. All patients were treated in our department with similar RT techniques and radiation doses. The demographic and clinical features of these patients are summarized in Table 1. No significant differences in clinical features were observed between the two groups. Of the 11 in-field recurrent cases, 7 were confirmed by biopsy, 3 by PET/CT, and 1 by CT. The sites of recurrence were the anastomosis in 5 patients, mediastinal lymph nodes in 5 patients, and left the main bronchus (invasion) in 1 patient.
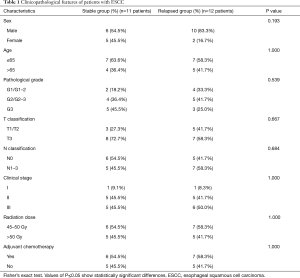
Full table
Dissimilarities in somatic mutation characteristics between the two groups
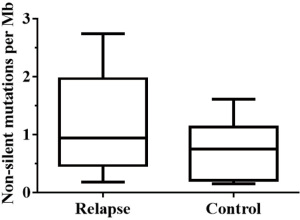
We achieved a read coverage of 145-fold in the 7 tissues from recurrent tumors and 23 pairs of tumor-normal samples. The average mutation rate was 0.95 nonsilent mutations per Mb; 75.1% of the genetic variants were SNVs, 13.7% were deletions, and 10.5% were insertions. For the 12 patients with relapsed ESCC, the median nonsilent mutation rate was 1.15 per Mb (range, 0.18–2.74 per Mb). For the 11 stable tumors, the nonsilent mutation rate per Mb was 0.73 (range, 0.30–1.61) (Figure S1). This nonsilent mutation rate was slightly lower than recently reported mutation rates in Chinese patients with ESCC (2.4–2.9 mutations per Mb) (12,18,19), possibly because many low-quality mutations were filtered out through of stringent parameter settings during this sequencing process.
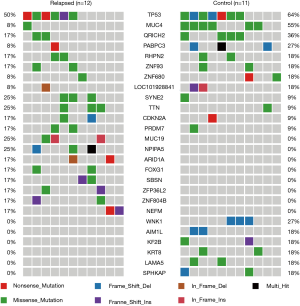
The mutation spectrum was compared between the two cohorts. Mutations in TP53, MUC4, and QR1CH2 were the most frequent alterations observed in this study (50%, 8%, and 17% and 64%, 55%, 36% in cohort 1 and cohort 2, respectively). In cohort 1, the relapsed ESCC group, the TP53 (50%), SYNE2 (25%), TTN (25%), MUC19 (25%), and NPIPA5 (25%) genes were significantly mutated, with at least three nonsilent mutations. Moreover, we found 392 genes that were unique to the relapsed group, and the spectrum showed the top 8 of these (MUC19, NPIPA5, ARID1A, FOXG1, SBSN, ZFP36L2, ZNF804B, and NEFM). In cohort 2, the stable ESCC group, TP53 (64%), MUC4 (55%), QR1CH2 (36%), RHPN2 (27%), and WNK1 (27%) were the significantly mutated genes. A total of 192 genes were unique to the stable group, and the spectrum showed the top 6 of these (WNK1, AIM1L, KIF2, KRT8, LAMA5, and SPHKAP) (Figure 1).
Survival analysis of the two groups
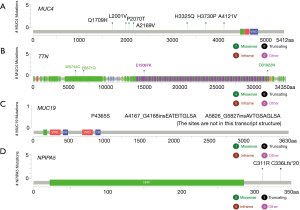
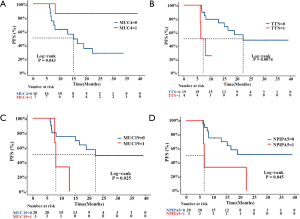
A total of 23 patients with ESCC were included in this study. In cohort 1, the relapsed ESCC group, all 12 ESCC patients, had new events, including death, after PORT. The median follow-up time for determining PFS was 8.3 months (range, 6.2–22.3 months; mean, 11.4 months). In cohort 2, the stable ESCC group, all 11 ESCC patients were confirmed to be in stable condition. The median follow-up time for determining PFS was undefined (range, 10.4–39.2 months; mean 23.8 months). According to the survival analysis, MUC4 [log-rank P=0.043; HR, 0.1592 (95% CI, 0.0491–0.5161)], TTN [log-rank P=0.0074; HR, 0.2088 (95% CI, 0.02105–2.07)], MUC19 (log-rank P=0.025; HR, 0.2591 (95% CI, 0.03201–2.098)] and NPIPA5 [log-rank P=0.045; HR, 0.2873 (95% CI, 0.03875–2.13)] gene mutations were significant prognostic and predictive indicators (Figure 2) in ESCC. The specific mutation sites in the MUC4, TTN, MUC19, and NPIPA5 genes are shown in Figure S2.
Copy number alterations (CNAs)
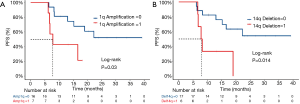
Somatic copy number alterations (SCNAs) can affect gene expression by altering gene dosage, disrupting coding sequences, and changing the genome structure. Many SCNAs are associated with various diseases (20-22). To investigate SCNAs in ESCC, we first detected chromosome arm-level events in tumor samples. Consequently, a copy number gain of 1q and deletion of 14q were observed to be associated with a risk of recurrence after postoperative chemoradiotherapy. The gain of 1q was detected in 7 (30.4%) of the 23 patients with ESCC: 2 in the stable group and 5 in the relapsed group. We observed a statistically significant difference in PFS time between patients with different 1q gain statuses: the median PFS time was 7.8 months for those with 1q gain and undefined for those without 1q gain [log-rank P=0.03; HR, 0.312 (95% CI, 0.07205–1.351)] (Figure 3A). Deletion of 14 was detected in 6 (26.1%) of the 23 patients with ESCC: 1 in the stable group and 5 in the relapsed group. We also observed a statistically significant difference in PFS time between patients with different 14q deletion statuses: the median PFS time was 8 months for those with deletion of 14q and undefined for those without deletion of 14q [log-rank P=0.014; HR, 0.2729 (95% CI, 0.05902–1.262)] (Figure 3B).
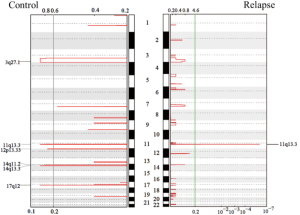
Furthermore, the GISTIC module was used to identify the significantly amplified or deleted regions of the genome in the 23 tumors analyzed by WES. A copy number gain of 11q13.3 was observed frequently in both groups, and gains of 3q27.1, 12p13.33, 14q11.2, 14q13.3, and 17q12 were observed in the stable group, while no deletions were observed in either group (Figure S3).
Discussion
To our knowledge, this is the first deep sequencing study comparing favorable-prognosis and unfavorable-prognosis patients with ESCC treated with PORT to identify radioresistance-related biomarkers in esophageal cancer. RT is one of the most important therapeutic strategies for EC. However, due to radioresistance, the proportion of infield recurrence after high dose RT may still be high, even for patients treated with PORT (23). For decades, this problem has been a significant challenge in radiation oncology. The heterogeneous nature of radioresistance has already been described by others (5,16,24). Finding predictive biomarkers is confusing because several factors may be associated with radioresistance. Therefore, new research design and advanced research technology are needed to address this issue.
In this study, using WES, we performed such an analysis of mutational features in two independent cohorts of ESCC patients who had received PORT and revealed the differences in the mutation spectrum between the two groups. We found that the mutation rate in the relapsed group was slightly higher than that in the stable group. The mutation types and proportions in the two groups did not differ significantly; C>T transition was the primary type, similar to reported mutation types in Chinese patients with ESCC (18). Furthermore, critical genetic differences were observed, and genetic alterations specific to the relapsed group were detected, including alterations in MUC4, TTN, MUC19, NPIPA5, ARID1A, FOXG1, SBSN, ZFP36L2, ZNF804B, and NEFM. Finally, three mutated genes (TTN, MUC19, and NPIPA5) and two CNAs (1q amplification and 14q deletion) were identified to be associated with poor RT prognosis, while MUC4 was a favorable factor.
MUC4 is membrane-bound mucin that promotes carcinogenic progression and is often proposed as a promising biomarker for various carcinomas (25-28). Acting as a ligand for the interaction with ErbB, MUC4 can bind HER2 and activate several downstream signaling proteins, including ERK1/2, Akt, FAK, and c-Src, among others (29-31). MUC4 is also found in both normal and ESCC tissues, and its expression correlates with all stages of squamous cell differentiation (32). Additionally, MUC4 has been reported to play a vital role in the proliferative and migratory properties of esophageal cancer cells (33). In this study, the MUC4 missense mutation rate in the stable ESCC group was noticeably higher than that in the relapsed group (55% vs. 8%), and patients with MUC4 missense mutations had much longer PFS times than those without these mutations (Figure 2). Several papers have reported that overexpression of MUC4 causes various treatment resistance phenotypes through diverse signaling pathways (27,34,35), including antiapoptotic activity and G0/G1 cell cycle arrest. The radiation response is also associated with apoptosis and the distribution of the cell cycle. Therefore, we speculated that MUC4 missense mutation was likely to weaken postoperative radioresistance in ESCC patients. However, the functional consequences of MUC4 missense mutation in ESCC and its corresponding roles in radioresistance remain to be elucidated.
TTN, also named cardiomyopathy, dilated 1G (CMD1G), the longest known coding gene, is emerging as a significant gene in human inherited disease, and its truncation mutations are strongly related to cardiac and skeletal muscle diseases (36). TTN has recently been reported to be mutated frequently in many tumor types, including lung squamous cell carcinoma, lung adenocarcinoma, and breast cancer (37-39). In this study, the TTN missense mutation rate in the relapsed ESCC group was noticeably higher than that in the stable group (25% vs. 9%), and patients without TTN missense mutations had much longer PFS times than those with mutations (Figure 2). Lin et al. (40) reported that lncRNA-TTN-AS1, derived from the antisense strand of the TTN gene, promoted ESCC cell proliferation and metastasis. Furthermore, cell cycle analysis confirmed ESCC cells overexpressing lncRNA-TTN-AS1 exhibited a significantly reduced G1 population, and a markedly increased S phase population. According to the radiobiology theory, S-phase cells are remarkably resistant to radiation. This phenomenon could explain why patients with TTN missense mutations had short PFS times. However, more research must explain the core relationship between TTN missense mutation and radioresistance.
Both MUC19 and NPIPA5 were identified to be associated with poor RT prognosis in this study. Furthermore, MUC19 was identified as a novel gland-specific gel-forming mucin gene and mainly expressed in the mucous cells of various glands, including the primary salivary glands (41). MUC19 mutation was reported in melanoma (42), and patients with high MUC19 expression exhibited worse prognosis in breast cancer (43). However, whether MUC19 participates in the tumorigenesis or radioresistance of ESCC is still unclear. NPIPA5 mutation has rarely been reported to be associated with human tumors, which might deserve further investigations to study the functional relevance of individual mutational events.
The ARID1A gene is usually described as a tumor suppressor, which recurrently mutated in many types of cancers, including ovarian, gastric, bladder, and breast cancers (44). ARID1A has also been reported as a novel tumor suppressor gene in Barrett’s esophagus pathogenesis (45). ARID1A actively participates in the regulation of several cellular events, including cell cycle arrest (46), DNA repair, and nonhomologous end-joining (NHEJ) activity (47). All these events are among the mechanisms that could lead to resistance to radiation and chemotherapy. In this study, the ARID1A mutation was unique to the recurrence group, although the mutation rate of ARID1A was not very high (17%), and a significant association with PFS was not found. Whether the ARID1A mutation is related to ESCC radioresistance, remain to be further studied. This research supplied a fundamental basis on ARID1A and ESCC.
Also, both 1q copy number gain and 14q deletion are identified to be associated with poor RT prognosis in this study. Many previous studies (48-50) have shown that a gain of 1q is a poor prognostic factor in various malignancies, including ESCC and esophageal adenocarcinoma (51,52). In particular, Merchant et al. (49) reported that patients with infratentorial tumors without 1q gain have higher event-free survival (EFS) rates with surgery and conformal radiation therapy than those with 1q gain. These results were like those in this study. Patients with 1q amplification have a poor prognosis after PORT, suggesting that 1q is indeed associated with radioresistance. Also, the importance of 14q in tumor suppression has been validated in esophageal cancer (53). The conclusions of these studies were consistent with that in this study. Considering the above results, one or more putative genes located on chromosome arms 1q and 14q may be associated with radioresistance.
Although this study has a small sample size and precludes us from reaching any firm conclusions about mutations associated with radioresistance in ESCC, we can draw several potentially essential conclusions. First, our study showed that comparing the gene mutation spectrum of patients with ESCC whose prognoses are opposite after PORT is a practical approach to study the genetic mechanisms of radioresistance, which are worthy of further exploration. Second, we found that MUC4, TTN, MUC19, NPIPA5, 1q, and 14q mutations were explicitly associated with ESCC radioresistance, and these findings can potentially be used to guide subsequent studies.
Additionally, mutations participate in heterotypic interactions with one another and interact with diverse signaling pathways and identify novel putative drivers and future directions for functional studies that are challenging and remain mostly unresolved. Thus, a more homogeneous study population with well-characterized clinical data is urgently needed.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5196
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-5196
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5196). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Approval from the Institutional Research Ethics Committee of Affiliated Hospital of Nantong University (Jiangsu, China) (No. 2019-K078) and prior consent from patients was obtained for using the clinical materials for research purposes. The study was performed following the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Deng W, Lin SH. Advances in radiotherapy for esophageal cancer. Ann Transl Med 2018;6:79. [Crossref] [PubMed]
- Li H, Fang W, Yu Z, et al. Chinese expert consensus on mediastinal lymph node dissection in esophagectomy for esophageal cancer (2017 edition). J Thorac Dis 2018;10:2481-9. [Crossref] [PubMed]
- Herskovic A, Martz K, al-Sarraf M, et al. Combined chemotherapy and radiotherapy compared with radiotherapy alone in patients with cancer of the esophagus. N Engl J Med 1992;326:1593-8. [Crossref] [PubMed]
- Krause M, Dubrovska A, Linge A, et al. Cancer stem cells: Radioresistance, prediction of radiotherapy outcome and specific targets for combined treatments. Adv Drug Deliv Rev 2017;109:63-73. [Crossref] [PubMed]
- Diehn M, Cho RW, Lobo NA, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 2009;458:780-3. [Crossref] [PubMed]
- Schulz A, Meyer F, Dubrovska A, et al. Cancer Stem Cells and Radioresistance: DNA Repair and Beyond. Cancers (Basel) 2019;11:862. [Crossref] [PubMed]
- Skvortsova I, Debbage P, Kumar V, et al. Radiation resistance: Cancer stem cells (CSCs) and their enigmatic pro-survival signaling. Semin Cancer Biol 2015;35:39-44. [Crossref] [PubMed]
- Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer 2011;11:393-410. [Crossref] [PubMed]
- Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nature Genetics 2010;42:30-5. [Crossref] [PubMed]
- Cheng C, Zhou Y, Li H, et al. Whole-Genome Sequencing Reveals Diverse Models of Structural Variations in Esophageal Squamous Cell Carcinoma. Am J Hum Genet 2016;98:256-74. [Crossref] [PubMed]
- Zhang L, Zhou Y, Cheng C, et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet 2015;96:597-611. [Crossref] [PubMed]
- Dulak AM, Stojanov P, Peng S, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet 2013;45:478-86. [Crossref] [PubMed]
- Deng J, Chen H, Zhou D, et al. Comparative genomic analysis of esophageal squamous cell carcinoma between Asian and Caucasian patient populations. Nat Commun 2017;8:1533. [Crossref] [PubMed]
- Cancer Genome Atlas Research N, Analysis Working Group: Asan U, Agency BCC, et al. Integrated genomic characterization of oesophageal carcinoma. Nature 2017;541:169-75. [Crossref] [PubMed]
- Baumann M, Krause M, Overgaard J, et al. Radiation oncology in the era of precision medicine. Nat Rev Cancer 2016;16:234-49. [Crossref] [PubMed]
- Amarasinghe KC, Li J, Hunter SM, et al. Inferring copy number and genotype in tumour exome data. BMC Genomics 2014;15:732. [Crossref] [PubMed]
- Gao YB, Chen ZL, Li JG, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet 2014;46:1097-102. [Crossref] [PubMed]
- Dai W, Ko JMY, Choi SSA, et al. Whole-exome sequencing reveals critical genes underlying metastasis in oesophageal squamous cell carcinoma. J Pathol 2017;242:500-10. [Crossref] [PubMed]
- Zhang L, Feizi N, Chi C, et al. Association Analysis of Somatic Copy Number Alteration Burden With Breast Cancer Survival. Front Genet 2018;9:421. [Crossref] [PubMed]
- Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet 2013;45:1134-40. [Crossref] [PubMed]
- Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010;463:899-905. [Crossref] [PubMed]
- Zhang W, Liu X, Xiao Z, et al. Postoperative intensity-modulated radiotherapy improved survival in lymph node-positive or stage III thoracic esophageal squamous cell carcinoma. Oncol Res Treat 2015;38:97-102. [Crossref] [PubMed]
- Barker HE, Paget JTE, Khan AA, et al. The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nature Reviews Cancer 2015;15:409-25. [Crossref] [PubMed]
- Jonckheere N, Van Seuningen I. Integrative analysis of the cancer genome atlas and cancer cell lines encyclopedia large-scale genomic databases: MUC4/MUC16/MUC20 signature is associated with poor survival in human carcinomas. J Transl Med 2018;16:259. [Crossref] [PubMed]
- Senapati S, Chaturvedi P, Chaney WG, et al. Novel INTeraction of MUC4 and galectin: potential pathobiological implications for metastasis in lethal pancreatic cancer. Clin Cancer Res 2011;17:267-74. [Crossref] [PubMed]
- Mercogliano MF, De Martino M, Venturutti L, et al. TNFα-Induced Mucin 4 Expression Elicits Trastuzumab Resistance in HER2-Positive Breast Cancer. Clin Cancer Res 2017;23:636-48. [Crossref] [PubMed]
- Kaur S, Sharma N, Krishn SR, et al. MUC4-mediated regulation of acute phase protein lipocalin 2 through HER2/AKT/NF-kappaB signaling in pancreatic cancer. Clin Cancer Res 2014;20:688-700. [Crossref] [PubMed]
- Bafna S, Kaur S, Batra SK. Membrane-bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene 2010;29:2893-904. [Crossref] [PubMed]
- Ponnusamy MP, Singh AP, Jain M, et al. MUC4 activates HER2 signalling and enhances the motility of human ovarian cancer cells. Br J Cancer 2008;99:520-6. [Crossref] [PubMed]
- Vincent A, Ducourouble MP, Van Seuningen I. Epigenetic regulation of the human mucin gene MUC4 in epithelial cancer cell lines involves both DNA methylation and histone modifications mediated by DNA methyltransferases and histone deacetylases. FASEB J 2008;22:3035-45. [Crossref] [PubMed]
- Guillem P, Billeret V, Buisine MP, et al. Mucin gene expression and cell differentiation in human normal, premalignant and malignant esophagus. Int J Cancer 2000;88:856-61. [Crossref] [PubMed]
- Bruyere E, Jonckheere N, Frenois F, et al. The MUC4 membrane-bound mucin regulates esophageal cancer cell proliferation and migration properties: Implication for S100A4 protein. Biochem Biophys Res Commun 2011;413:325-9. [Crossref] [PubMed]
- Macha MA, Rachagani S, Pai P, et al. MUC4 regulates cellular senescence in head and neck squamous cell carcinoma through p16/Rb pathway. Oncogene 2015;34:1698-708. [Crossref] [PubMed]
- Reynolds IS, Fichtner M, McNamara DA, et al. Mucin glycoproteins block apoptosis; promote invasion, proliferation, and migration; and cause chemoresistance through diverse pathways in epithelial cancers. Cancer Metastasis Rev 2019;38:237-57. [Crossref] [PubMed]
- Ware JS, Amor-Salamanca A, Tayal U, et al. Genetic Etiology for Alcohol-Induced Cardiac Toxicity. J Am Coll Cardiol 2018;71:2293-302. [Crossref] [PubMed]
- Cheng X, Yin H, Fu J, et al. Aggregate analysis based on TCGA: TTN missense mutation correlates with favorable prognosis in lung squamous cell carcinoma. J Cancer Res Clin Oncol 2019;145:1027-35. [Crossref] [PubMed]
- Jia Y, Duan Y, Liu T, et al. LncRNA TTN-AS1 promotes migration, invasion, and epithelial mesenchymal transition of lung adenocarcinoma via sponging miR-142-5p to regulate CDK5. Cell Death Dis 2019;10:573. [Crossref] [PubMed]
- Lips EH, Michaut M, Hoogstraat M, et al. Next generation sequencing of triple negative breast cancer to find predictors for chemotherapy response. Breast Cancer Research 2015;17:134. [Crossref] [PubMed]
- Lin C, Zhang S, Wang Y, et al. Functional Role of a Novel Long Noncoding RNA TTN-AS1 in Esophageal Squamous Cell Carcinoma Progression and Metastasis. Clin Cancer Res 2018;24:486-98. [Crossref] [PubMed]
- Chen Y, Zhao YH, Kalaslavadi TB, et al. Genome-wide search and identification of a novel gel-forming mucin MUC19/Muc19 in glandular tissues. Am J Respir Cell Mol Biol 2004;30:155-65. [Crossref] [PubMed]
- Cifola I, Pietrelli A, Consolandi C, et al. Comprehensive genomic characterization of cutaneous malignant melanoma cell lines derived from metastatic lesions by whole-exome sequencing and SNP array profiling. PLoS One 2013;8:e63597. [Crossref] [PubMed]
- Song L, Xiao Y. Downregulation of hsa_circ_0007534 suppresses breast cancer cell proliferation and invasion by targeting miR-593/MUC19 signal pathway. Biochem Biophys Res Commun 2018;503:2603-10. [Crossref] [PubMed]
- Kadoch C, Hargreaves DC, Hodges C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 2013;45:592-601. [Crossref] [PubMed]
- Streppel MM, Lata S, DelaBastide M, et al. Next-generation sequencing of endoscopic biopsies identifies ARID1A as a tumor-suppressor gene in Barrett's esophagus. Oncogene 2014;33:347-57. [Crossref] [PubMed]
- Nagl NG Jr, Patsialou A, Haines DS, et al. The p270 (ARID1A/SMARCF1) subunit of mammalian SWI/SNF-related complexes is essential for normal cell cycle arrest. Cancer Res 2005;65:9236-44. [Crossref] [PubMed]
- Watanabe R, Ui A, Kanno S, et al. SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res 2014;74:2465-75. [Crossref] [PubMed]
- Chagtai T, Zill C, Dainese L, et al. Gain of 1q As a Prognostic Biomarker in Wilms Tumors (WTs) Treated With Preoperative Chemotherapy in the International Society of Paediatric Oncology (SIOP) WT 2001 Trial: A SIOP Renal Tumours Biology Consortium Study. J Clin Oncol 2016;34:3195-203. [Crossref] [PubMed]
- Merchant TE, Bendel AE, Sabin ND, et al. Conformal Radiation Therapy for Pediatric Ependymoma, Chemotherapy for Incompletely Resected Ependymoma, and Observation for Completely Resected, Supratentorial Ependymoma. J Clin Oncol 2019;37:974-83. [Crossref] [PubMed]
- Chiang J, Dalton J, Upadhyaya SA, et al. Chromosome arm 1q gain is an adverse prognostic factor in localized and diffuse leptomeningeal glioneuronal tumors with BRAF gene fusion and 1p deletion. Acta Neuropathol 2019;137:179-81. [Crossref] [PubMed]
- van Dekken H, Wink JC, Vissers KJ, et al. Genomic analysis of early adenocarcinoma of the esophagus or gastroesophageal junction: tumor progression is associated with alteration of 1q and 8p sequences. Genes Chromosomes Cancer 2006;45:516-25. [Crossref] [PubMed]
- Cui XB, Tian YX, Chun CP, et al. Genome-wide screening for genomic aberrations in Kazakh patients with esophageal squamous cell cancer by comparative genomic hybridization. Int J Clin Exp Pathol 2018;11:427-37. [PubMed]
- Ko JM, Yau WL, Chan PL, et al. Functional evidence of decreased tumorigenicity associated with monochromosome transfer of chromosome 14 in esophageal cancer and the mapping of tumor-suppressive regions to 14q32. Genes Chromosomes Cancer 2005;43:284-93. [Crossref] [PubMed]
(English Language Editor: J. Chapnick)


