Genetic alterations in cfDNA of benign and malignant thyroid nodules based on amplicon-based next-generation sequencing
Introduction
Thyroid nodules are extremely common. About 4–7% of the population develop palpable thyroid nodules (1), while 19–68% of people have nodules detected incidentally by thyroid ultrasonography (2). In 8–16% of cases, thyroid nodules are malignant and require surgery (1), with papillary thyroid carcinoma (PTC) comprising 90% of these cases (3). Therefore, distinguishing between malignant and benign nodules is clinically important. Currently, ultrasound-guided fine needle aspiration cytology (FNAC) is the gold standard for differentiating benign nodules from malignant ones, as recommended by European and American guidelines (2,4). Nevertheless, in approximately 20–30% of cases, the lesions are non-diagnostic and indeterminate or suspicious (5), with about 34% of these eventually revealed to be malignant (6,7). In addition, FNA can only investigate one lesion at a time and is an invasive procedure that even when performed by experienced doctors still carries a risk of postoperative bleeding.
The American Thyroid Association (ATA) guidelines for patients with thyroid nodules (2) suggested that in patients with indeterminate FNA cytology, a seven-gene panel of genetic mutations and rearrangements (BRAF, RAS, RET/PTC, PAX8/PPARγ), a gene expression classifier (GEC) of 167 genes and galectin 3 immunohistochemistry can provide molecular diagnostic assessment. In addition, TERT promoter mutations were found in 7–22% of PTC and more than 70% of anaplastic thyroid carcinoma (ATC), and TP53 was present in more than 70% of ATC (3). Currently, it is believed that if a BRAF, TERT, or TP53 mutation exists or a fusion gene is detected, thyroid cancer is almost present (8,9). Recent studies have shown the potential role of miRNA markers in the diagnosis of indeterminate FNA specimens (10,11), but it has not been fully verified. In addition to single focal papillary thyroid microcarcinoma (PTMC), PTC takes thyroidectomy and lymph node dissection as the primary treatment. In some patients, radioactive iodine (RAI) treatment and TSH suppression play an adjunct role. Serum Tg assays and neck ultrasound are the main contents of PTC follow-up (2,4). In order to avoid overtreatment, it has been proposed to use the seven-gene panel in patients with indeterminate FNA cytology as the basis for decision-making on primary thyroid surgery (12), but there is insufficient evidence at present (2). Currently predictive value of molecular marker is still limited for distinguishing benign and malignant thyroid nodules and belong to invasive diagnostic method. Thus, considerable effort has been made to identify other reliable markers for primary PTC.
First discovered in 1994, the detection of circulating cell-free DNA (cfDNA) serves as a non-invasive liquid biopsy (13). Numerous studies have shown that an elevated level of cfDNA is reflective of the pathogenesis of stroke, trauma, inflammatory disease, sepsis, and especially malignant diseases (14). In malignant diseases, the release of cfDNA is related to the apoptosis and necrosis of cancer cells in the tumor microenvironment and is secreted by cancer cells (14,15). Analysis of cfDNA assists with the detection of tumor-related genetic alterations and avoids the need for tumor tissue biopsy. Moreover, it is useful for the implementation of precision medicine. Circulating tumor DNA (ctDNA) was recognized in one prospective, single-center study to be a specific and highly sensitive biomarker of metastatic breast cancer (16). In addition, cfDNA has been indicated to potentially play a role in the non-invasive diagnosis and monitoring of malignant tumors such as pancreatic cancer, non-small cell lung cancer (NSCLC), and melanoma (17-19). Another study (20) of 24 patients with KRAS wild-type colorectal cancer before monotherapy with panitumumab (a therapeutic anti-EGFR antibody) showed that 9 patients were found to have KRAS mutation in their serum during treatment, indicating that cfDNA detection can be used to guide clinical treatment programs.
Many previous studies have used qPCR or high-sensitivity PCR to detect changes in cfDNA of PTC patients (21-24). However, the role of cfDNA detection in the diagnosis and monitoring of PTC is still controversial. Whether genetic alterations, such as BRAFV600E mutation, in cfDNA can be used as a biomarker for the diagnosis of thyroid carcinoma, or whether differential expression levels of cfDNA exist between benign and malignant thyroid nodules, is currently unknown. Recent studies have shown that next-generation sequencing (NGS) based on amplicons has obvious advantages in the detection of cfDNA (25). Therefore, in this study, DNA mutations in tissue, plasma, and blood cell samples from patients with PTC and nodule goiter were detected using amplicon-based NGS to analyze the differential expression of cfDNA in benign and malignant thyroid nodules, with the aim of exploring potential biomarkers for clinical diagnosis.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-4544).
Methods
Patient samples
Twenty patients with thyroid nodules, including 10 patients with benign nodular goiter and 10 patients with PTC, who were treated in the First Affiliated Hospital of Sun Yat-sen University (Guangzhou, China) were enrolled in our study from July 2017 to November 2017. The inclusion criteria are (I) at least 18 years old; (II) PTC or nodular goiter diagnosed by histology or cytology; (III) the first discovered PTC or nodular goiter; (IV) for the first time to undergo thyroid surgery. The exclusion criteria are (I) patients with other malignant tumors; (II) those who have received chemotherapy for antithyroid cancer treatment or radioactive iodine treatment; (III) those with severe liver and kidney dysfunction; (IV) disagree to sign the informed consent form of this study. The diagnoses of all of the patients were confirmed by postoperative pathology. Three sample types were collected from each patient: tumor tissue [formalin-fixed and paraffin-embedded (FFPE) sections], blood cells, and plasma. The samples were obtained along with informed consent from each patient. The study was approved by the Institutional Research Ethics Committee of Sun Yat-sen University. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Sample processing
The FFPE sections of thyroid tumor tissue were 5–8 µm thick. DNA was extracted from one or two sections from each sample. Blood samples (5 mL) were collected in EDTA tubes and centrifuged at 1,900 rpm for 10 min within 2 h of sample collection. Plasma and blood cells were then separately collected and stored at −80 and −20 °C, respectively.
DNA isolation and purification
Genomic DNA was obtained from the FFPE, blood cell, or plasma samples using QIAamp DNA FFPE Tissue Kit (QIAGEN, Dusseldorf, Germany, Cat No. 56404), QIAamp DNA Blood Mini Kit (QIAGEN, Cat No. 51106), and QIAamp Circulating Nucleic Acid Kit (QIAGEN, Cat No. 55114), respectively, according to the manufacturer’s instructions. Isolated DNA was homogeneously mixed, and stored at −20 °C. The concentrations of DNA samples and the libraries prepared were determined using a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). Then, 10 ng of each DNA product was analyzed by capillary electrophoresis on a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA).
Primer design
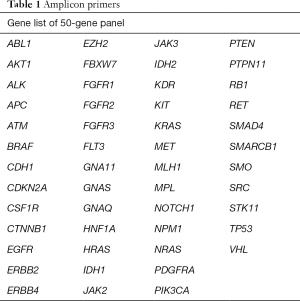
To cover exons with hot spot mutations, 207 pairs of amplicon primers were designed from 50 proto-oncogenes and tumor suppressor genes using AmpliSeqTM (Lifetech, Inc., USA) (Table 1). Amplicon libraries were generated with amplicons ranging from 91 to 137 bp. Designs were made to accommodate the nature of small DNA fragments of both FFPE samples and blood samples.
Full table
Amplicon library preparation
The amplicon library was prepared using DNA Seq Library Preparation Kit for Amplicon Sequencing-Illumina Compatible (Gnomegene, San Diego, CA, USA, K02422A-L). The library preparation workflow comprised the five straightforward steps, starting with genomic DNA isolated from FFPE, blood cells, and plasma samples.
Amplicons were amplified using 10 ng genomic DNA from blood cells and FFPE samples for 20 PCR cycles and different initial quantities cfDNA for 23/25 PCR cycles. Subsequently, end repair, adapter ligation, and a simple PCR amplification were performed in line with the manufacturer’s instructions with several purification steps to obtain library products with different indexes (Gnomegene, K02422A-L). The PCR conditions consisted of an initial denaturation step for 45 sec at 98 °C followed by several cycles of denaturation for 10 sec at 98 °C, annealing for 30 sec at 60 °C and extension for 30 sec at 72 °C with a final incubation of 72 °C for 7 min. The number of PCR cycles was adjusted according to the quantity of sample. Size selection was carried out as the last step to obtain clean library products. Then, 10 ng of each library product was analyzed by capillary electrophoresis with a Bioanalyzer 2100 (Agilent) to confirm that the expected fragment size had been obtained and to determine the concentration. Following the size selection and quality control, purified and quantified libraries were mixed for each run. All library products were diluted to 26 pM in molecular biology grade water.
Illumina sequencing
Different samples require different depths of sequencing. Libraries prepared from DNA samples from FFPE or blood cell need a sequencing depth of 1,000×, whereas libraries prepared from cfDNA samples need a sequencing depth of 10,000×. The constructed library was sequenced using Hiseq X10 PE 150 (Illumina, San Diego, California, USA). After sequencing, signal processing and base calling was performed.
SNP calling and filtering
Raw sequencing data reads of all 60 samples, which contained reads sequence and corresponding sequencing quality, were acquired from sequencers. After standardized quality control procedures, clean sequencing data reads were produced. The clean sequencing data reads were then mapped on human reference genome (version GRCh37.p13) with BWA MEM (26). To avoid mapping error triggered by repeat sequences, only uniquely mapped reads were kept.
Detection of somatic mutation
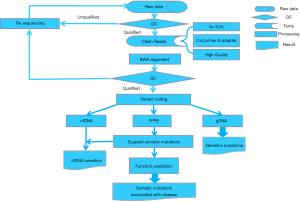
After quality control and some routine data processing, first, we detected the mutations for each sample with varScan (version v2.3). The options for mutation detection were: (I) base quality value ≥20; (II) coverage ≥50×; (III) number of reads supporting mutation ≥5; (IV) no chain specificity; (V) two test P value ≤0.05; (VI) mutation frequency ≥1%. Then, the somatic mutations were identified by comparing the mutations detected in the blood cell samples and the FFPE samples (Figure 1).
Results
Clinical characteristics and sequencing data
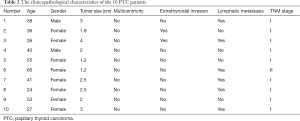
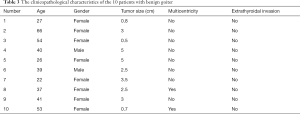
A total of 20 patients were included in our research, including 10 patients with benign nodular goiter and 10 patients with PTC. The PTC and nodular goiter patients had an average age of 40.6 and 40.5 years, respectively. Both groups had a male to female ratio of 1:4. According to the TNM staging system, 9 PTC patients were classified as stage I and 1 as stage II (Tables 2 and 3).
Full table
Full table
Primers for hot spot mutations were designed from 50 proto-oncogenes and tumor suppressor genes (Table 1). Amplicon sequencing was subsequently used to detect gene mutations in DNA extracted from a total of 60 tissue, blood cell, and plasma samples from 20 patients. The align rate and the ratio of the target area of all these 60 samples were about 85%. The sequencing depth of the plasma samples was around 6,000×, and those of the blood cell and FFPE samples were around 2,000×. Sequencing specificity and coverage uniformity were about 85% and 90%, respectively.
Somatic mutations in benign and malignant thyroid nodules
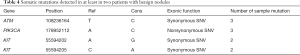
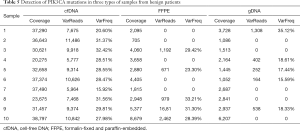
To analyze the somatic mutations in thyroid nodules, mutations in tissues were detected and compared with genetic mutations in blood cells. Table 4 shows the somatic mutations detected in at least two patients with benign thyroid tumors, one of which was a non-synonymous mutation. The mutation was subsequently detected in the plasma, tissue, and blood cell samples of the 10 patients with benign thyroid tumors (Table 5). The results showed that the mutation also had a high frequency in the other five blood cell samples. Therefore, we speculate it may not be a somatic mutation.
Full table
Full table
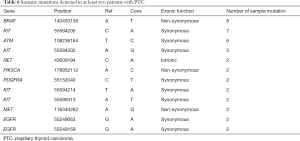
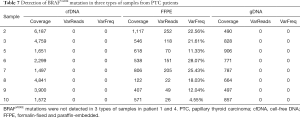
Twelve somatic mutations were detected in at least two patients with malignant thyroid tumors (Table 6), three of which were non-synonymous mutations. We further analyzed these three mutations in the three sample types from the 10 patients with malignant thyroid tumors (Table 7; Tables S1 and S2). The BRAFV600E gene mutation was detectable in the tissue samples of eight PTC patients, indicating that BRAFV600E mutation is a very credible disease-related mutation.
Full table
Full table

Full table
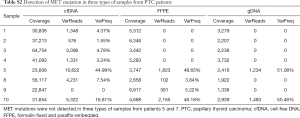
Full table
Differential analysis in cfDNA between benign and malignant nodules
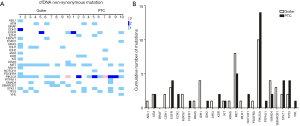
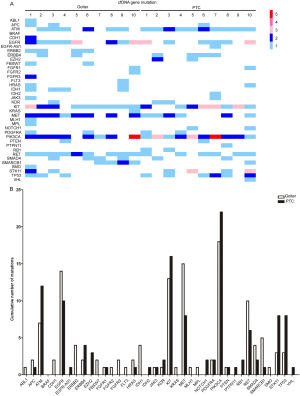
The BRAFV600E mutation was detected in FFPE samples from patients with malignant tumors, but it did not appear in all of the samples from the benign patients, suggesting that it could serve as a reliable marker to distinguish malignant tumors from benign tumors. However, in our study, the mutation was not detected in cfDNA samples from patients with malignant tumors (Figure 2A; Figure S1A). To distinguish cfDNA in malignant and benign samples, we analyzed the two sets of cfDNA samples to find the difference between them (Figure 2A,B; Figure S1). There was no significant difference in the number of mutations in cfDNA between the benign and malignant patients. In relation to the distribution of mutation genes, there was a large difference between the two sets of cfDNA samples, and some genes had more mutations in benign patients than in malignant patients, including MET and IDH1. Some genes had more mutations in malignant patients than in benign patients, such as PIK3CA and EZH2.
Discussion
There is currently no non-invasive method for definitive diagnosis of thyroid carcinoma. As the incidence of thyroid carcinoma in the population increases (27), non-invasive diagnostic method becomes more needed. As a non-invasive diagnostic tool, cfDNA can effectively avoid the risks that FNA may bring. Because of its repeatability, it can also replace Tg as a means of monitoring tumor dynamics. Moreover, in patients with postoperative tumor recurrence, cfDNA can be a potential diagnostic tool when tissue is not easy to obtain.
In thyroid carcinoma, common genetic alterations include BRAFV600E mutation, RAS mutation, PTEN deletion or mutation, PIK3CA amplification or mutation, RET/PTC fusion, and PAX8/PPARγ rearrangement (28). Based on previous studies, BRAFV600E mutation in PTC has a global incidence of about 45% (28), which is as high as 85% in China (29-31). BRAFV600E mutation in PTC is related factors associated with poor prognosis such as extrathyroidal invasion, lymph node metastasis, late TNM staging, 131I treatment resistance, and recurrent disease (32,33). In our study, similarly, we found that in the tissue sequencing results of 10 PTC patients, BRAFV600E mutation was present in 8, although it could not be detected in the cfDNA of most of the PTC patients.
The current study is still in conflict about the detection of BRAFV600E mutation in serum.
Cradic et al. (34) used an allele-specific real-time PCR, which can detect BRAFV600E mutation in PTC patients with a detection sensitivity of fewer than one heterozygote BRAFV600E-carrying cell per 100,000 diploid cells, and determined that BRAFV600E mutation could be detected in the serum of 20 out of 173 PTC patients. Among 42 patients whose tissues were BRAFV600E mutation positive, the blood of 8 (19%) patients was also BRAFV600E positive. Another study from Pupilli’s group (23) pointed out that by using a similar method, the percentage of BRAFV600E mutation in blood was correlated to cytological type, among which PTC was the highest. The results also showed a lower percentage of BRAFV600E in cfDNA from a second sample blood taken after, rather than before, surgery. Chuang et al. (35), using the gap-ligase chain reaction technique, could detect 3 BRAFV600E mutations (60%) in the serum of 5 PTC patients with BRAFV600E-positive tissue samples. Of the five patients, two had lymph node metastases, two had Hashimoto’s thyroiditis, and one had distant metastasis. Using peptide nucleic acid (PNA) clamp real-time PCR, Kim et al. (22) found that 3 (6.1%) of 49 patients with PTC tumors with BRAFV600E mutation tested positive for BRAFV600E mutation in both their plasma and tumor tissue; these 3 PTC patients had lymph node and lung metastases.
However, several studies (24,36,37) that used qRT-PCR, and even extremely sensitive digital PCR, found that the identification of BRAFV600E mutation is not possible in the cfDNA of PTC patients, even in those who have distant metastasis with BRAFV600E mutation-positive tumor tissue.
PCR technology applied to cfDNA can only detect one or more genes to monitor the development of disease or therapeutic response. The detection of dozens or even hundreds of hotspot gene mutations depends on sequencing technology. Therefore, in this study, we used amplicon-based NGS technology. Several previous studies have verified the sensitivity of this sequencing technology for the detection of cfDNA in various tumor plasmas. Based on this sequencing technology, Guibert’s group (38) found that the sensitivity of detecting EGFR mutation in the cfDNA of patients with non-small cell lung cancer was 100%, compared with 87% for droplet digital PCR (ddPCR), and even rare driver oncogenic mutations could be detected. Page’s research (39), which focused on metastatic breast cancer, used an amplicon panel covering 16 genes and found that half of patients had at least one mutation or amplification in cfDNA, while no mutations were identified in the cfDNA of the healthy controls. Osumi’s study (25) indicated that somatic mutations in cfDNA were detectable in 88 of 101 patients with metastatic colorectal cancer, of which the KRAS mutation rate was 38.6% and the concordance rate between cfDNA and matched tissue was 77.2%. Therefore, researchers believed that the amplicon-based NGS system could sensitively detect mutant alleles of cfDNA.
In our research, we were not able to detect BRAFV600E mutation in the cfDNA of PTC patients. This may be explained by the study’s insufficient sample size. The differential expression of cfDNA caused by the individual differences in PTC patients may also be a potential reason for the large gap between our results and those of similar studies. Furthermore, compared with previous research, the difference in detection accuracy between different detection methods may have also influenced the results. In addition, the proportion of cfDNA that originates from tumor cells varies depending on the stage and size of the tumor (14). Most of the samples included in our study were from stage I/II patients, which may have resulted in low cfDNA concentration and increased difficulty in detection. Some previous studies have also failed to detect BRAFV600E mutation in the cfDNA of PTC patients with distant metastases (24). The smaller tumor volume of PTC and the relatively lower shedding of cfDNA in PTC may be the reason for the low detection rate of cfDNA in PTC patients.
Previous research also indicated that an elevated concentration of cfDNA can occur in benign lesions, inflammation, and trauma (14). In our study, sequencing was also used to detect abnormal cfDNA expression in benign thyroid nodules. Although no mutation was found as a biomarker, we found that the number of cfDNA mutations in benign nodules was not significantly different from that in malignant nodules as with previous research (40); however, their mutation gene profiles were different. Genes such as MET and IDH1 mutated more frequently in benign nodules than in malignant nodules. These results indicate that it may be possible to use differential gene profiles in cfDNA to identify benign and malignant lesions through multi-gene joint risk assessment.
Cell free nucleic acid (cfNA) includes DNA, RNA, and miRNA. Although cfDNA may not be especially useful as a biomarker in this sequencing result, the epigenetic alterations, besides genetic alterations, may be more meaningful for detection. Our previous research (41) found that compared with healthy controls, the expression of let-7e, miR-151-5p, and miR-222 in the serum of PTC patients was significantly increased. In addition, there was a series of studies which described the expression profile of miRNA in PTC serum (42). Furthermore, some studies have discovered that the expression of Runx2 mRNA in the serum and circulating non-hematopoietic cells of PTC patients was higher than that in normal donors, and that it was positively correlated with the expression of Runx2 mRNA in puncture specimens (43). The measurement of circulating thyroglobulin-specific transcript mRNA level was used to predict the possibility of early detection of tumor recurrence (44). The findings of these studies suggest that the detection of epigenetic alterations may also hold great potential as a biomarker for diagnosis and recurrence monitoring of PTC patients.
Conclusions
In conclusion, amplicon sequencing was used to detect hotspot mutations in benign and malignant thyroid nodules, and BRAFV600E mutation was found to be a credible disease-related mutation in PTC; however, it could not be detected in cfDNA. The results also revealed a large difference in mutation gene distribution between benign and malignant thyroid nodules.
Acknowledgments
Funding: This research was supported by grants from the National Natural Science Foundation of China (No. 81772850 and No. 81701367).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-4544
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-4544
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4544). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Research Ethics Committee of Sun Yat-sen University. The informed consent was obtained from each patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Burman KD, Wartofsky L. Clinical practice. Thyroid Nodules. N Engl J Med 2015;373:2347-56. [Crossref] [PubMed]
- Haugen BR, Alexander EK, Bible KC, et al. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: The American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer. Thyroid 2016;26:1-133. [Crossref] [PubMed]
- Fagin JA, Wells SJ. Biologic and Clinical Perspectives on Thyroid Cancer. N Engl J Med 2016;375:1054-67. [Crossref] [PubMed]
- Filetti S, Durante C, Hartl D, et al. Thyroid cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann Oncol 2019;30:1856-83. [Crossref] [PubMed]
- Kiernan CM, Solorzano CC. Bethesda Category III, IV, and V Thyroid Nodules: Can Nodule Size Help Predict Malignancy? J Am Coll Surg 2017;225:77-82. [Crossref] [PubMed]
- Wang CC, Friedman L, Kennedy GC, et al. A large multicenter correlation study of thyroid nodule cytopathology and histopathology. Thyroid 2011;21:243-51. [Crossref] [PubMed]
- Cibas ES, Ali SZ. The 2017 Bethesda System for Reporting Thyroid Cytopathology. Thyroid 2017;27:1341-6. [Crossref] [PubMed]
- Durante C, Grani G, Lamartina L, et al. The Diagnosis and Management of Thyroid Nodules: A Review. JAMA 2018;319:914-24. [Crossref] [PubMed]
- Eszlinger M, Lau L, Ghaznavi S, et al. Molecular profiling of thyroid nodule fine-needle aspiration cytology. Nat Rev Endocrinol 2017;13:415-24. [Crossref] [PubMed]
- Shen R, Liyanarachchi S, Li W, et al. MicroRNA signature in thyroid fine needle aspiration cytology applied to "atypia of undetermined significance" cases. Thyroid 2012;22:9-16. [Crossref] [PubMed]
- Agretti P, Ferrarini E, Rago T, et al. MicroRNA expression profile helps to distinguish benign nodules from papillary thyroid carcinomas starting from cells of fine-needle aspiration. Eur J Endocrinol 2012;167:393-400. [PubMed]
- Nikiforov YE, Ohori NP, Hodak SP, et al. Impact of mutational testing on the diagnosis and management of patients with cytologically indeterminate thyroid nodules: a prospective analysis of 1056 FNA samples. J Clin Endocrinol Metab 2011;96:3390-7. [Crossref] [PubMed]
- Vasioukhin V, Anker P, Maurice P, et al. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br J Haematol 1994;86:774-9. [Crossref] [PubMed]
- Schwarzenbach H, Hoon DSB, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [Crossref] [PubMed]
- Choi JJ, Reich CR, Pisetsky DS. The role of macrophages in the in vitro generation of extracellular DNA from apoptotic and necrotic cells. Immunology 2005;115:55-62. [Crossref] [PubMed]
- Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199-209. [Crossref] [PubMed]
- Liu X, Liu L, Ji Y, et al. Enrichment of short mutant cell-free DNA fragments enhanced detection of pancreatic cancer. EBioMedicine 2019;41:345-56. [Crossref] [PubMed]
- Murtaza M, Dawson S, Tsui DWY, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [Crossref] [PubMed]
- Valpione S, Gremel G, Mundra P, et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur J Cancer 2018;88:1-9. [Crossref] [PubMed]
- Diaz LA Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012;486:537-40. [Crossref] [PubMed]
- Salvianti F, Giuliani C, Petrone L, et al. Integrity and Quantity of Total Cell-Free DNA in the Diagnosis of Thyroid Cancer: Correlation with Cytological Classification. Int J Mol Sci 2017;18:1350. [Crossref] [PubMed]
- Kim BH, Kim IJ, Lee BJ, et al. Detection of plasma BRAF(V600E) mutation is associated with lung metastasis in papillary thyroid carcinomas. Yonsei Med J 2015;56:634-40. [Crossref] [PubMed]
- Pupilli C, Pinzani P, Salvianti F, et al. Circulating BRAFV600E in the diagnosis and follow-up of differentiated papillary thyroid carcinoma. J Clin Endocrinol Metab 2013;98:3359-65. [Crossref] [PubMed]
- Condello V, Macerola E, Ugolini C, et al. Analysis of circulating tumor DNA does not improve the clinical management of patients with locally advanced and metastatic papillary thyroid carcinoma. Head Neck 2018;40:1752-8. [Crossref] [PubMed]
- Osumi H, Shinozaki E, Takeda Y, et al. Clinical relevance of circulating tumor DNA assessed through deep sequencing in patients with metastatic colorectal cancer. Cancer Med 2019;8:408-17. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [Crossref] [PubMed]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer 2013;13:184-99. [Crossref] [PubMed]
- Guo L, Ma YQ, Yao Y, et al. Role of ultrasonographic features and quantified BRAFV600E mutation in lymph node metastasis in Chinese patients with papillary thyroid carcinoma. Sci Rep 2019;9:75. [Crossref] [PubMed]
- Li XJ, Mao XD, Chen GF, et al. High BRAFV600E mutation frequency in Chinese patients with papillary thyroid carcinoma increases diagnostic efficacy in cytologically indeterminate thyroid nodules. Medicine (Baltimore) 2019;98:e16343. [PubMed]
- Yan C, Huang M, Li X, Wang T, Ling R. Relationship between BRAF V600E and clinical features in papillary thyroid carcinoma. Endocr Connect 2019;8:988-96. [Crossref] [PubMed]
- Xing M, Alzahrani AS, Carson KA, et al. Association between BRAF V600E mutation and recurrence of papillary thyroid cancer. J Clin Oncol 2015;33:42-50. [Crossref] [PubMed]
- Xing M, Alzahrani AS, Carson KA, et al. Association between BRAF V600E mutation and mortality in patients with papillary thyroid cancer. JAMA 2013;309:1493-501. [Crossref] [PubMed]
- Cradic KW, Milosevic D, Rosenberg AM, et al. Mutant BRAF(T1799A) can be detected in the blood of papillary thyroid carcinoma patients and correlates with disease status. J Clin Endocrinol Metab 2009;94:5001-9. [Crossref] [PubMed]
- Chuang TC, Chuang AY, Poeta L, et al. Detectable BRAF mutation in serum DNA samples from patients with papillary thyroid carcinomas. Head Neck 2010;32:229-34. [PubMed]
- Zane M, Agostini M, Enzo MV, et al. Circulating cell-free DNA, SLC5A8 and SLC26A4 hypermethylation, BRAF(V600E): A non-invasive tool panel for early detection of thyroid cancer. Biomed Pharmacother 2013;67:723-30. [Crossref] [PubMed]
- Kwak JY, Jeong JJ, Kang SW, et al. Study of peripheral BRAF(V600E) mutation as a possible novel marker for papillary thyroid carcinomas. Head Neck 2013;35:1630-3. [Crossref] [PubMed]
- Guibert N, Hu Y, Feeney N, et al. Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non-small cell lung cancer. Ann Oncol 2018;29:1049-55. [Crossref] [PubMed]
- Page K, Guttery DS, Fernandez-Garcia D, et al. Next Generation Sequencing of Circulating Cell-Free DNA for Evaluating Mutations and Gene Amplification in Metastatic Breast Cancer. Clin Chem 2017;63:532-41. [PubMed]
- Lupo M, Guttler R, Geck Z, et al. Is measurement of circulating tumor dna of diagnostic use in patients with thyroid nodules? Endocrine Practice 2018;24:453-9. [Crossref] [PubMed]
- Yu S, Liu Y, Wang J, et al. Circulating microRNA profiles as potential biomarkers for diagnosis of papillary thyroid carcinoma. J Clin Endocrinol Metab 2012;97:2084-92. [Crossref] [PubMed]
- Perdas E, Stawski R, Nowak D, et al. The Role of miRNA in Papillary Thyroid Cancer in the Context of miRNA Let-7 Family. Int J Mol Sci 2016;17:909. [Crossref] [PubMed]
- Dalle Carbonare L, Frigo A, Francia G, et al. Runx2 mRNA expression in the tissue, serum, and circulating non-hematopoietic cells of patients with thyroid cancer. J Clin Endocrinol Metab 2012;97:E1249-56. [Crossref] [PubMed]
- Barzon L, Boscaro M, Pacenti M, et al. Evaluation of circulating thyroid-specific transcripts as markers of thyroid cancer relapse. Int J Cancer 2004;110:914-20. [Crossref] [PubMed]