GLA missense and promoter variants co-segregating in a Chinese family with Fabry disease
Introduction
Fabry disease (FD; MIM 301500) is an X-linked lysosomal storage disorder caused by mutations in the GLA gene leading to deficient α-galactosidase A (α-Gal A) activity, glycosphingolipid accumulation, and life-threatening complications. There is wide clinical variability of the different mutations, including anhidrosis, angiokeratoma, corneal dystrophy, recurrent episodes of neuropathic pain, renal impairment, cardiac complications and cerebrovascular manifestations. FD is of interest to specialties across the spectrum of internal medicine but is notoriously difficult to diagnose. From the onset of symptoms to a definitive diagnosis, the recognition of the underlying diagnosis is delayed by 14 years in males and 19 years in females (1).
Whole-exome sequencing (WES) technology has become very popular for the diagnosis of rare genetic diseases that have been confusing in the clinic (2). The GLA gene is the only gene found to be currently related to FD (3). WES technology can not only identify the pathogenic genes of single gene genetic diseases but also has superior suggestive significance for the diagnosis of potential polygenic diseases.
There is a certain variability between the genotype and the clinical phenotype in FD. Patients with FD who have the same genotype have different clinical manifestations, and some patients do not have FD but have the GLA gene mutations (4,5). Therefore, the study of the relationship between genotype and phenotype, and the variant gene function is important for genetic counseling, diagnosis and prognosis.
Here, we performed WES on affected members of a four generation Chinese Han kindred with Fabry disease and filtered the data to identify coding and non-coding variants in GLA. Male patients have more severe symptoms than female patients. We present the following article in accordance with the STREGA reporting checklist (available at http://dx.doi.org/10.21037/atm-19-4510).
Methods
Pedigree and subjects
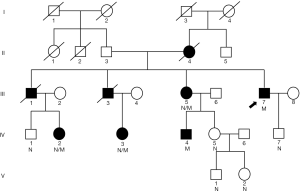
A 5-generation, 26-member Chinese Han family with FD from Shandong Province, China, was recruited from the Chinese PLA General Hospital (Figure 1). Ten family members of the pedigree were involved in this study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) which was approved by Medicine Ethics Committee of Chinese PLA General Hospital. The informed consent was taken from all the patients.
Clinical test
All 10 individuals underwent skin and ophthalmic examinations, urinalysis, serum creatinine tests, and echocardiogram. Patient history of neuropathic pain, hypohidrosis or anhidrosis was also collected. The proband underwent skin and kidney biopsies.
Whole-exome sequencing
For next-generation sequencing and SNP genotypes, genomic DNA was extracted from participants’ blood samples using the Qiagen DNA Blood Mini Kit (QIAGEN Bioinformatics). A target region of 62,085,286 bp (2.00% of the genome) (Agilent Technologies Inc.) was captured and sequenced using an Hiseq X10 instrument (Illumina). The average coverage was 100X. Paired-end reads of 150 bp were generated. Reads were aligned to the UCSC hg19 human reference genome using Burrows-Wheeler Aligner BWA-MEM (http://biobwa.sourceforge.net/).
Exome capture, sequencing and alignment of the target DNA were performed using Illumina TruSeq.
Quality score recalibration was performed during alignment, and reads that aligned to multiple locations were discarded. Following alignment, presumed PCR duplicates were removed using MarkDuplicates.jar from Picard.
Bioinformatics Analysis of the Mutation
WES genotype inference and linkage analysis of SNP genotypes were inferred from WES data using the SAMtools mpileup and bcftools view commands from release 916 of the SAMtools package, which infers genotypes using a revised version of the MAQ SNP model. We required base quality and mapping quality to be ≥13. SAMtools produces a variant call format (VCF) file, from which we extracted genotypes using a Perl script.
Illumina bcl2fastq2 was used to generate fastq files. All sequencing reads were aligned to the human genome reference sequence (UCSC hg19) using the BWA-MEM algorithm. Picard tools were used to mark/remove PCR duplicates and to calculate sequencing metrics. The point mutations and indels were called using both GATK4 and commercial software (Sentieon). Functional annotation of variants was performed using ANNOVAR, which contains more than 40 databases, such as 1000g2014, ExAC, esp6500, etc.
Functional prediction tools, including Polymorphism Phenotyping version 2 (PolyPhen-2) SIFT, and MutationTaster, were used to evaluate the possible effects of amino acid alteration on protein structure and function.
Data analysis for bioinformatics annotation after WES
(I) Functional regions of mutation sites: only analysis of exon regions, splice site regions, and intron or UTR regions reported in the literature; (II) type of mutation: filter out synonymous mutations that have not been reported in any literature; (III) frequency of 1,000-genomes: analysis of mutation sites below 1% of 1000g2014, and the other reference database: ExAC, esp6500; (IV) screening for genes related to clinical symptoms based on clinical symptom analysis; (V) this case is a family sample, and targeted analysis of mutation sites based on the incidence of the family; (VI) analyze the pathogenicity of the mutation sites that selected finally according to Mendel’s genetic law.
Functional study of g.1170C >T mutation
Nine hundred and seventy-two bps wild type GLA promoter sequence were cloned into pGL4-luc2 (Promega. Madison, USA) between Kpn I and Nco I site using PCR, named pGL4-GAL-WT. also we obtained the mutation vector named pGL4-GAL-MT included the mutation site ATGCTGTCCGGTTACCGTGACA. Two plasmids were transfected into 293T cell individually (Promega. Madison, USA), and hRluc vector, expresses Rellina, was used as the internal control. Cells were collected and assay with the dual luciferase kit (E1910, Promega. Madison, USA).
Statistical analyses
Quantitative data are expressed as means ± SDs. Statistical analysis was conducted using the GraphPad Prism software. One-way ANOVA were used to compare the differences of two groups. P<0.05 was considered significantly different.
Results
Clinical data
Seven people (II4, III1, III3, III5, III7, IV3 and IV4) from three generations exhibited different clinical symptoms; the main manifestations were neuropathic pain, angiokeratoma, and renal impairment. Three of the seven people were dead. Two were the proband’s brothers, and one was his mother. His two brothers (III1, III3) died of renal failure at 42 and 34 years of age, accompanied by neuropathic pain and angiokeratoma. His mother (II4), who died of natural causes, only manifested neuropathic pain and angiokeratoma. A retrospective study found that their clinical manifestations were consistent with the classical symptoms of FD. Of the 5 surviving symptomatic subjects (III5, III7, IV2, IV3, and IV4), 2 men (III7 and IV4) had kidney damage, and 2 women (III5, IV2, and IV3) showed only neuropathic pain, hypohidrosis or angiokeratoma. The 10 individuals included all 5 living symptomatic persons in the family. Medical history, laboratory tests, and WES was performed for 10 of the family members (The results of clinical data are summarized in Table 1). Proband (III7) is the oldest, at 47 years old. The diagnosis of FD was made based on typical histological features from renal biopsy. Skin biopsy also suggests typical angiokeratoma. The youngest is only 2 years old (V1).
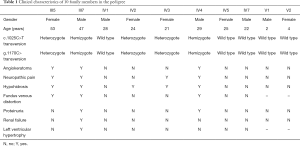
Full table
Pathological results of renal biopsy
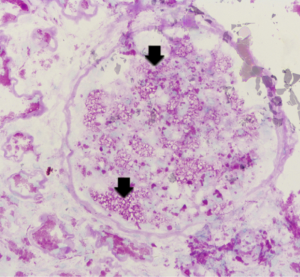
Renal biopsy results from the proband (III7) revealed vague vacuolization of podocytes on microscopic examination after staining by periodic acid-Schiff (PAS) (Figure 2). Section lining epithelial cell proliferation and wall segment thickening were also observed. The renal tubular epithelial cells showed granular, vacuolar, and foamy changes (Figure 3). There was an infiltration of renal interstitial inflammatory cells, severely fibrosis, and internal renal artery wall thickening.
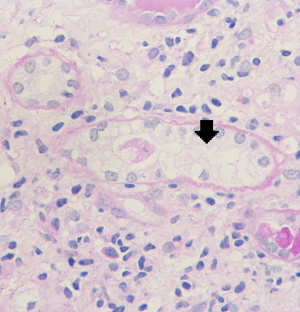
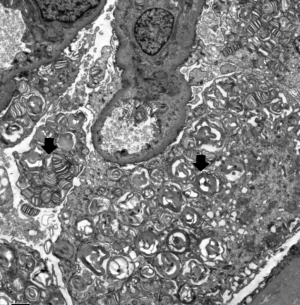
Electron microscopy revealed obvious vacuolar degeneration of capillary endothelial cells, typical electron-dense multilamellar inclusions and zebra bodies and an increase in secondary lysosomes in the cytoplasm of epithelial cells (Figure 4).
Analysis of whole-exome sequencing data
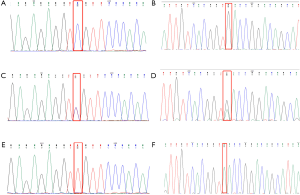
A total of 1,375 variants were found among 10 individuals by the whole-exome sequencing. Sanger sequencing verified that the GLA gene has a mutation c.1025C>T (p.Arg342Gln) (6-8) and g.1170C>T SNP (9) (Figure 5). Ten family members of the pedigree were involved in this study, including five affected individuals (III:5, III:7, IV:2, IV:3, and IV:4) and five unaffected individuals (IV:1, IV:5, IV:7, V:1 and V:2). Among the members with the c.1025C>T (p.Arg342Gln) mutation, 3 were female heterozygotes and 2 were male hemizygotes. All five also carried a g.1170C>T SNP in cis. The pedigree conforms to the typical characteristics of x-linked disorder. All five affected individuals from two generations in this family had two simultaneous variants in cis, c.1025C>T (p.Arg342Gln) and g.1170C>T, in the GLA gene. All of them had common clinical manifestations of FD, such as neuropathic pain and hypohidrosis. Especially male individuals, showed more severe clinical manifestations of FD, such as angiokeratoma, fundus venous distortion, and Proteinuria. Our data indicate that the c.1025C>T (p.Arg342Gln) mutation in the GLA gene was the disease-causing mutation in the family. g.1170C>T SNP occur in noncoding regions of the 5' untranslated region (UTR) of the human α-Gal gene but also have pathogenicity (9).
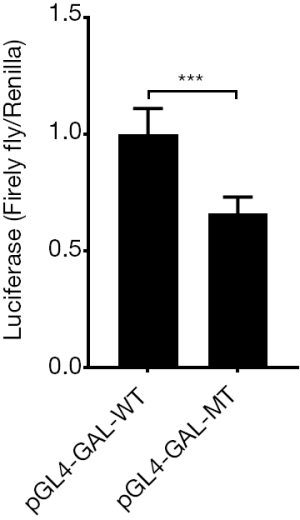
g.1170C>T SNP decrease the transcription of GLA gene
SNP in the GAL promoter, g.1170C>T, together with the side sequence formed to a potential transcriptional start site (TSS)—GTCACC. Therefore, we cloned the promoter of GAL including the TSS into the dual luciferase reporter system whose original Kozak site was deleted by Nco I restriction endonuclease. Vectors were transfected into the 293T cell. The assay results showed firefly/Rellina luciferase signal ratio in pGL4-GAL-MT group is weaker than WT group (0.58±0.10 vs. 1.00±0.18, P<0.005, Figure 6), it indicated the mutation of GAL DNA sequence would decrease the transcription of firefly gene after it.
Discussion
FD is a disease in which a series of mutations on the X-chromosome lead to defects in α-Gal enzyme production, resulting in decreased enzyme activity or synthesis effects and loss of enzyme activity. Accumulation of globo cyanuric ceramide (Gb3) and other sphingolipids in lysosomes in various tissues—mainly cardiac, endothelial, neuronal and renal—results in end-organ damage and failure (10).
This study is consistent with the observation that most female FD patients have less serious symptoms (11). The heterozygotes with inactivated healthy allele were inclined to have notably higher clinical severity score than women with random inactivation of both X-chromosomes. The skewed X-inactivation may also be responsible for clinical manifestations in female carriers of X-linked diseases. Normal heterozygotes could present skewed X-inactivation in favour of the normal allele. Disease manifestations in female heterozygotes are usually mild and have a slower rate of progression. Approximately 30% have minimal angiokeratomas, 19% have spiral appearance of fundus vessels (77% in male) (12), and less than 10% have infrequent attacks of neuropathic pains (13), which may be partly due to the result of lyonization (14). There were also phenotypic differences between male members in this family. Hemizygote male members have severe clinical symptoms and wild-type male members have mild clinical symptoms. It was affected by gene mutations, and also related to gene expression. It may be affected by the individual ’s external environment such as age, other diseases, and other genetic variations .The two GLA gene variants in cis found in this study, c.1025C>T (p.Arg342Gln) and g.1170C>T, occurred simultaneously in the 5 affected individuals, in which the c.1025C>T (p.Arg342Gln) mutation occurred in the coding region, and g.1170C>T SNP occurred in the noncoding region (5'UTR).The mutation affects transcription of GLA gene were also confirmed by dual luciferase reporter system.
The c.1025C>T (p.Arg342Gln) mutation was first reported in Dutch Caucasians with FD in 1994 and was discovered by direct sequencing of the GLA gene (15). A single G-A transition in exon 7 leads to an arginine-to-glutamine (Arg342Gln) missense mutation, but it has not been studied in a typical family. The Arg342Gln mutation is associated with α-Gal A deficiency, which was not through changes in protein secondary structure. Arginine and glutamine are both able to establish hydrogen bonding, but whereas glutamine can form only one hydrogen bond, arginine can form two hydrogen bonds. This extra hydrogen bond of arginine can be of major importance for the conservation of the tertiary structure or may play a role in substrate binding (15).
In 2008, g.1170C>T was first reported in the Portuguese population. It is one of three SNPs in the 5'UTR of exon 1 (9), located on the X chromosome (chrX: 100662901), 10 base pairs upstream of the GLA gene, which is a point mutation in the promoter. The g.1170T allele is associated with lower a-Gal expression. As mRNA 5'UTR sequences are not translated, the most plausible mechanism(s) underlying the impact of the g.1170C>T SNP on a-Gal expression would operate at transcriptional or translational control (16,17). Oliveira et al. indicated that the g.1170C>T SNP may be co-dominantly associated with a relatively decreased GLA expression at the transcription and/or translation level (18). The dual luciferase experiment is required for further verification (19). To confirm this, we demonstrated that the g.1170C>T SNP can decline subsequent transcription of the GLA gene using the dual luciferase reporter system, presumably the transcription start site of the gene. More detailed functional verification needs to be studied in the future using EMSA and other experiments.
Two variants in cis in the GLA gene were rare reported. Two missense mutations were found associated in the same allele in a French patient with the typical form: D313Y + G411D (c.937G4T + c.1232G4A) (20). But expression studies are needed to demonstrate if they are both Fabry disease causing mutations as in other reported patients with two associated missense mutations. Mao et al. reported a double mutation: the frameshift mutation c.273_276del TGAT (p.I90MfsTer25) with the missense mutation (c.281G>T, p.Cys94Phe) in three-generation Chinese pedigree using whole exon sequencing. Familial episodic pain using was the only clinical manifestation of the affected members (21). A Japanese patient with two combined mutations 66Glu→Gln (196G→C)/112Arg→Cys (334C→T) in exon 2 showed continuous shooting pain (22). Two unrelated classically FD males were each found to have two missense mutations: R112C + D313Y (c.334C4T + c.937G4T) and C172G + D313Y (c.514T4G + c.937G4T). Intracellular experiment indicated that pR112C/D313Y and pD313Y/G411D all had essentially no α-Gal A activity (23). An unbalanced expression of these two transcripts due to NM_000169.2:c.639+861C>T (g.9273C>T or IVS4+861C>T) and NM_000169.2:c.639+919G>A (g.9331G>A or IVS4+919G>A) GLA deep intronic mutations has been reported to cause FD with cardiac variant in Italian families (24,25). The clinical significance of the two variants in cis in FD is still not very clear.
The kidney phenotype of a two variants in cis male seemed more serious than the single-site mutation when compared to the clinical phenotype of a previously reported single mutation. A Portuguese Caucasian young adult male’s clinical symptoms only showed premature ischaemic small-vessel cerebrovascular disease (ISVCD) without renal involvement, whose only genomic variation found was the SNP (18). A 30-year-old man with FD only presented with angiokeratoma without kidney damage. The patient was shown to be hemizygote for a missense mutation g.1170C>T only in the GLA (6). While the main clinical symptom of the proband in our study was renal insufficiency. We speculated that double mutations may have a greater impact on protein structure and function than single mutations, which requires further computer simulation of protein structure and function.
This report shows an important point regarding kidney biopsy. The kidney is one of the main target organs of FD, and renal pathological examination has been widely carried out. Although the proband’s kidney specimen demonstrated a mildly abnormal appearance under light microscopy examination, electron microscopy clearly demonstrated lamellated myelin structures and confirmed the diagnosis of typical FD (26). If the characteristic changes of renal pathology can be fully understood, when they are combined with clinical multisystem performance, the diagnosis rate of FD can be greatly improved. Some clinically normal heterozygous females have typical kidney biopsy findings (27). GLA activity when measured in blood may not reflect the true level of the enzyme activity in the affected organs. Therefore the critical threshold of GLA activity is hard to be determined (28). This implies that biopsy of the involved organ could help us identify the extent of FD and start therapy accordingly.
With the maturity of next-generation sequencing technology, targeted capture sequencing, represented by whole-exome sequencing, provides a new method for FD diagnosis with multiple clinical phenotypes (29). In this study, high-throughput sequencing was used to further explore new possible pathogenic genes of FD in addition to the GLA gene by performing WES on the family to find more new mutation sites. The next-generation sequencing results found several other abnormal genes related to kidney, eye, heart and other symptoms, but they were not consistent with the family symptoms, and no sanger sequencing was performed. Copy number variants (CNVs) could be another type of genetic defect underlying FD that has not been investigated. The pathological/histological investigation was only done for the proband. Renal/skin biopsies from the other affected individuals can be very informative for correlating all three aspects (genetic/clinical and histological findings), but unfortunately we didn’t get permission from other individuals. The family members did not do the α-Gal activity test. Although enzyme replacement therapy (ERT) became widely available in 2001, there are no approved marketable drugs in China, so the proband has not undergone ERT (30).
Conclusions
In summary, we confirmed FD using WES. What we found for the first time was that c.1025C>T and g.1170C>T variants occurred at the same time. in the GLA gene in a Chinese Han family with FD. The pathogenicity of the variants and the relationship between genotype and phenotype were also confirmed. The mutations affects transcription of GLA gene, presumably the transcription start site by functional study. Renal biopsy characteristic osmiophilic inclusion bodies in electron microscopy also confirmed the pathogenicity. The interaction and functional studies, as well as in vivo and in vitro studies of genetically deficient animals, are warranted to facilitate a better understanding of the pathogenesis and development of targeted therapy.
Acknowledgments
We thank the participating patients and investigators for their cooperation and efforts in collecting the genetic information and DNA specimens.
Funding: This study was supported by The National Key Research and Development Program of China (No. 2016YFC1305503), Natural Science Foundation of China (Nos. 81700629, 81870491, 81830019).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at http://dx.doi.org/10.21037/atm-19-4510
Peer Review File: Available at http://dx.doi.org/10.21037/atm-19-4510
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-19-4510
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-19-4510). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) which was approved by Medicine Ethics Committee of Chinese PLA General Hospital. The informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Eng CM, Fletcher J, Wilcox WR, et al. Fabry disease: Baseline medical characteristics of a cohort of 1765 males and females in the Fabry Registry. J Inherit Metab Dis 2007;30:184-92. [Crossref] [PubMed]
- Cimmaruta C, Citro V, Andreotti G, et al. Challenging popular tools for the annotation of genetic variations with a real case, pathogenic mutations of lysosomal alpha-galactosidase. BMC Bioinformatics 2018;19:433. [Crossref] [PubMed]
- Bishop DF, Calhoun DH, Bernstein HS, et al. Human alpha-galactosidase A: nucleotide sequence of a cDNA clone encoding the mature enzyme. Proc Natl Acad Sci U S A 1986;83:4859-63. [Crossref] [PubMed]
- van der Tol L, Smid BE, Poorthuis BJ, et al. A systematic review on screening for Fabry disease: prevalence of individuals with genetic variants of unknown significance. J Med Genet 2014;51:1-9. [Crossref] [PubMed]
- Hasholt L, Ballegaard M, Bundgaard H, et al. The D313Y variant in the GLA gene - no evidence of a pathogenic role in Fabry disease. Scand J Clin Lab Invest 2017;77:617-21. [Crossref] [PubMed]
- Germain DP. Co-occurrence and contribution of Fabry disease and Klippel-Trenaunay-Weber syndrome to a patient with atypical skin lesions. Clin Genet 2001;60:63-7. [Crossref] [PubMed]
- Pasqualim G, Simon L, Sperb-Ludwig F, et al. Fabry disease: a new approach for the screening of females in high-risk groups. Clin Biochem 2014;47:657-62. [Crossref] [PubMed]
- Turaca LT, Pessoa JG, Motta FL, et al. New mutations in the GLA gene in Brazilian families with Fabry disease. J Hum Genet 2012;57:347-51. [Crossref] [PubMed]
- Oliveira JP, Ferreira S, Barcelo J, et al. Effect of single-nucleotide polymorphisms of the 5' untranslated region of the human alpha-galactosidase gene on enzyme activity, and their frequencies in Portuguese caucasians. J Inherit Metab Dis 2008;31 Suppl 2:S247-53. [Crossref] [PubMed]
- Cairns T, Muntze J, Gernert J, et al. Hot topics in Fabry disease. Postgrad Med J 2018;94:709-13. [Crossref] [PubMed]
- Dobrovolny R, Dvorakova L, Ledvinova J, et al. Relationship between X-inactivation and clinical involvement in Fabry heterozygotes. Eleven novel mutations in the alpha-galactosidase A gene in the Czech and Slovak population. J Mol Med (Berl) 2005;83:647-54. [Crossref] [PubMed]
- Fiore B, Klopfer M, Schwebig C, et al. Ocular motility disorders in a patient with Fabry's disease. Ophthalmologe 2009;106:544-6. [Crossref] [PubMed]
- MacDermot KD, Holmes A, Miners AH. Anderson-Fabry disease: clinical manifestations and impact of disease in a cohort of 60 obligate carrier females. J Med Genet 2001;38:769-75. [Crossref] [PubMed]
- Guo Y, Yuan J, Liang H, et al. Identification of a novel COL4A5 mutation in a Chinese family with X-linked Alport syndrome using exome sequencing. Mol Biol Rep 2014;41:3631-5. [Crossref] [PubMed]
- Ploos van Amstel JK, Jansen RP, de Jong JG, et al. Six novel mutations in the alpha-galactosidase A gene in families with Fabry disease. Hum Mol Genet 1994;3:503-5. [Crossref] [PubMed]
- Mignone F, Grillo G, Licciulli F, et al. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res 2005;33:D141-6. [Crossref] [PubMed]
- Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5'- and 3'-UTR-binding factors. Trends Biochem Sci 2003;28:182-8. [Crossref] [PubMed]
- Oliveira JP, Ferreira S, Reguenga C, et al. The g.1170C>T polymorphism of the 5' untranslated region of the human alpha-galactosidase gene is associated with decreased enzyme expression--evidence from a family study. J Inherit Metab Dis 2008;31 Suppl 2:S405-13. [Crossref] [PubMed]
- Kadam PD, Chuan HH. Erratum to: Rectocutaneous fistula with transmigration of the suture: a rare delayed complication of vault fixation with the sacrospinous ligament. Int Urogynecol J 2016;27:505. [Crossref] [PubMed]
- Guffon N, Froissart R, Chevalier-Porst F, et al. Mutation analysis in 11 French patients with Fabry disease. Hum Mutat 1998.Suppl 1:S288-90. [Crossref] [PubMed]
- Mao C, Luo H, Yang J, et al. Pseudo-dominant inheritance of a novel double GLA mutation associated with Fabry disease mimicking familial episodic pain. Am J Med Genet A 2016;170:3051-3. [Crossref] [PubMed]
- Ishii S, Sakuraba H, Suzuki Y. Point mutations in the upstream region of the alpha-galactosidase A gene exon 6 in an atypical variant of Fabry disease. Hum Genet 1992;89:29-32. [Crossref] [PubMed]
- Yasuda M, Shabbeer J, Benson SD, et al. Fabry disease: characterization of alpha-galactosidase A double mutations and the D313Y plasma enzyme pseudodeficiency allele. Hum Mutat 2003;22:486-92. [Crossref] [PubMed]
- Filoni C, Caciotti A, Carraresi L, et al. Unbalanced GLA mRNAs ratio quantified by real-time PCR in Fabry patients' fibroblasts results in Fabry disease. Eur J Hum Genet 2008;16:1311-7. [Crossref] [PubMed]
- Ishii S, Nakao S, Minamikawa-Tachino R, et al. Alternative splicing in the alpha-galactosidase A gene: increased exon inclusion results in the Fabry cardiac phenotype. Am J Hum Genet 2002;70:994-1002. [Crossref] [PubMed]
- Chong Y, Kim M, Koh ES, et al. Identification of a novel GLA mutation (Y88C) in a Korean family with Fabry nephropathy: a case report. BMC Med Genet 2016;17:76. [Crossref] [PubMed]
- Wuthrich RP, Weinreich T, Binswanger U, et al. Should living related kidney transplantation be considered for patients with renal failure due to Fabry's disease? Nephrol Dial Transplant 1998;13:2934-6. [Crossref] [PubMed]
- Schiffmann R, Fuller M. Is it Fabry disease? Genet Med 2016;18:1181-5. [Crossref] [PubMed]
- Peng H, Xu X, Zhang L, et al. GLA variation p.E66Q identified as the genetic etiology of Fabry disease using exome sequencing. Gene 2016;575:363-7. [Crossref] [PubMed]
- Biegstraaten M, Arngrimsson R, Barbey F, et al. Recommendations for initiation and cessation of enzyme replacement therapy in patients with Fabry disease: the European Fabry Working Group consensus document. Orphanet J Rare Dis 2015;10:36. [Crossref] [PubMed]