
The emerging prospects of circular RNA in tumor immunity
Introduction
More than 70% of genes in the human genome can be actively transcribed. However, only 1–2% of genes possess protein-coding ability, and most transcripts are non-coding RNA (ncRNA) (1). Circular RNA (circRNA) is a ncRNA commonly found in eukaryotes, which has abundance, stability, conservation, and a certain degree of tissue space specificity (2). circRNA has been found to be involved in the regulation of eukaryotic gene expression in various ways, including through binding to microRNA (miRNA) or RNA binding protein (RBP), interacting with transcription factors, encoding protein or polypeptide, and affecting pre-mRNA formation via competitive co-transcription (3-7). Moreover, the dysfunction of circRNA is associated with various diseases, including many types of tumors (8-13).
The importance of tumor immune response in tumor development is widely acknowledged (14). The earliest study on the response of the immune system to tumors used Coley’s toxins to trigger an immune response and treat a variety of inoperable cancers (15). In recent years, research on tumor immunity has become increasingly intensive. The immune system carries out a dual role in the promotion and suppression of tumorigenesis and tumor development (16). At the same time, the role of linear ncRNAs, such as miRNAs and long non-coding RNAs (lncRNAs), in tumor immunity has become clearer (17). However, the functions of circRNA in tumor immunity need further investigation. Herein, we review the existing research on circRNA in tumor immunity, with a focus on the promising role of circRNA in the regulation of tumor immunity.
The formation and characteristics of circRNA
In recent years, circRNA, a naturally occurring non-coding RNA, has received widespread attention. circRNA does not have a 5' cap or a 3' tail but forms a covalently closed circular structure, making it resistant to ribonuclease digestion with higher stability than linear mRNA (18).
circRNA formation
Unlike the canonical splicing of linear mRNA, circRNA is produced by non-canonical forms of alternative splicing. Based on various biosynthesis models, circRNAs are divided into three main categories: exonic circRNA (ecircRNA) (2), retained intron circRNA (ElciRNA) (19), and circular intronic RNA (ciRNA) (3) (Figure 1). Of these categories, ecircRNA is the largest in number, accounting for more than 80% of identified circRNAs. There are two primary modes of ecircRNA formation: lariat-driven circularization and intron-pair-driven circularization (2). Wilusz et al. proposed that circRNA is predominantly produced during alternative back-splicing, in which the splice donor site of the primary transcript’s downstream exon is connected to the splice acceptor site of the upstream exon in reverse order, forming a circular transcript. Moreover, exon circularization depends on the alternative pairing of flanking complementary intron sequences and inverted repeat Alu sequences, to enable the same gene locus to produce multiple circRNAs (20). Normally, introns between exons are spliced, but at some point, they are kept to form so-called retained-intron circRNAs (ElciRNAs) (5). Finally, in the nucleus of specific tissues, introns can independently circulate to form a circular intron RNA (ciRNA). ciRNA formation depends on a common motif containing a 7-nucleotide GU-rich element near the 5' splicing junction site and an 11-nucleotid C-rich element near the 3' splicing junction site, which allows the intron to avoid debranching and form a stable circRNA (3).
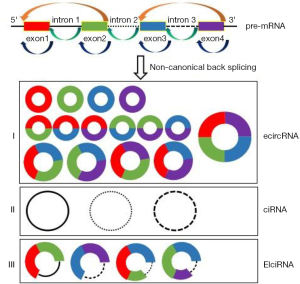
circRNA biosynthesis also depends on RNA-binding protein (RBP). Adenosine deaminase 1 (ADAR1), for example, cuts the formation of circRNA by binding the double-stranded RNA to the stem-loop structure (21). circRNA synthesis is inhibited by nuclear RNA helicase DHX9 through its explicit binding of the inverted repeat Alu sequence (IRAlus) (22). In contrast, circRNA formation is promoted by the splicing factors Quaking (QKI) (23) and Muscleblind (MBL) (7) as they bind to flanking introns of the circularized exons in their host gene mRNA precursors. The above studies show that RBP may play a vital role in the circularization of circRNA by inhibiting established splicing and bridging complementary sequences. However, the detailed mechanism of circRNA biogenesis needs further exploration.
Characteristics of circRNA
The prominent features of circRNA are as follows: (I) abundance: although the expression abundance of circRNA varies in different cells (24), the content of circRNA in some cells can be as high as ten fold more than its corresponding linear transcript (2). (II) Stability: the covalently closed loop structure of circRNA facilitates its resistance to exonuclease RNase R, meaning that circRNA is more stable than linear mRNA. Data has shown that in most species, the average half-life of circRNA exceeds 48 h, while the half-life of mRNA is about 10 h (25). (III) Conservation: most circRNAs have highly conserved sequences, meaning circRNAs are conserved among distinct species. For example, many circRNAs can be simultaneously detected in humans, mice, and even fruit flies (2,26). (IV) Distribution: most of the exon-derived circRNAs (ecircRNAs) are primarily located in cytoplasm and usually function as miRNA response elements (MRE) (27,28). A small number of intron-derived circRNAs, ciRNAs, and EIciRNAs, are located in the nucleus and may be involved in the regulation of gene expression at the transcriptional or post-transcriptional level (2,3). (V) Tissue and space specificity: the expression level of circRNA in the mammalian brain, for example, is high, especially at the synaptic site, and circRNA is usually upregulated during neuronal differentiation (29).
The functions of circRNA
Based on the above characteristics of circRNAs, a growing body of evidence has revealed that circRNAs might take part in the regulation of eukaryotic gene expression by exerting a series of functions. Herein, we enumerated several prominent biological functions of circRNAs (Figure 2).
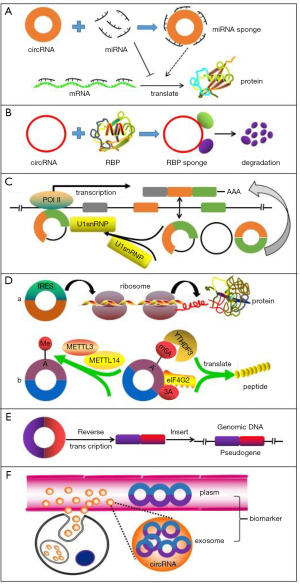
circRNA as miRNA sponge
Some circRNAs contain miRNA binding sites, which could serve as competitive endogenous RNA (ceRNA) to compete for miRNA binding sites to regulate miRNA activity negatively, thereby reducing the inhibitory effect of miRNA on its downstream target genes (30). One of the most typical examples is the antisense transcript of cerebellar degeneration-related protein-1 (CDR1as), also known as ciRS-7, which contains 74 selectively conserved miRNA binding sites and can serve as a molecular sponge for miR-7 (30). CDR1as can be cleaved in the nucleus by binding to miR-671 at the miR-7 target in an AGO2-dependent manner site, before miR-7 is transported to the subcellular site and released via the promotion of miR-671 (30). Studies have confirmed that ciRS-7 overexpression increases the expression of miRNA target genes, while knockdown of ciRS-7 has the opposite effect (30). Moreover, mouse testis-specific circRNA sex-determining region Y (circSry) has 16 miR-138 target sites and is associated with testicular development (30). circHIPK2 has been reported to be a molecular sponge for miR124-2HG, which regulates the activation of astrocytes through the synergistic effect of autophagy and endoplasmic reticulum (ER) stress (31). circBIRC6 regulates pluripotency and differentiation of human embryonic stem cells (hESCs) by binding to miR-34a and miR-145 (32). Other studies have found that circHIPK3 can function as a sponge for miR-124, which can upregulate the expression of the target genes IL6R and DXL2 of miR-124 through the ceRNA mechanism and promote cell proliferation in liver cancer (33). However, notably, the expression of most circRNAs in mammals is low, and the existence of multiple binding sites for the same miRNA is rare (34).
circRNA as RBP sponge
Studies have shown that circRNAs can bind to RBPs such as QKI, AGO, and MBL, to play an essential role in tumor progression as RBP sponges (35,36). CircRNA can bind to QKI protein in EMT and participate in the regulation of tumor metastasis (23). QKI-5, as a widely studied RBP, is considered as a novel tumor suppressor in many cancers, including lung and prostate cancer (37,38). The abnormal overexpression of AGO proteins has also been found in cancers and is closely related to the development of cancers that depend on miRNA-dependent or non-dependent pathways (39). These studies show that circRNA may interact with RBP and play an essential role in tumorigenesis and tumor development. Besides, circ-Foxo3 binds to cyclin-dependent kinase 2 (CDK2) and p21 to form a Foxo3-p21-CDK2 ternary complex that inhibits CDK2 function and blocks the cell cycle process, thereby participating in the regulation of cancer development (40). circPABPN1 has also been reported to inhibit the binding of HuR to PABPN1 mRNA by binding to HuR, which provides the first example of the competition between circRNA and its homologous mRNA for RBP (41).
circRNA can regulate transcription and splicing
Although most circRNA is found in the cytoplasm, EIciRNA and ciRNA retained in the nucleus may be involved in transcriptional regulation. Studies have revealed that by knocking out several EIciRNAs, the transcription of their parent genes can be reduced. EIciRNA can interact with U1 small nuclear ribonucleoprotein (U1snRNP), and the EIciRNA-U1 snRNP complex interacts with RNA polymerase II (Pol II) on the promoter of its parent gene to enhance gene expression. Blocking this RNA-RNA interaction disrupts the interaction between EIciRNA and Pol II and later reduces the transcription of its parent gene. For example, circEIF3J and circPAIP2 interact with U1 snRNP and Pol II to enhance the transcription of their parent genes (5). circSEP3 is a retaining nuclear circRNA derived from exon 6 of SEPALLATA3 (SEP3) in Arabidopsis thaliana, which regulates the splicing of linear transcripts. circSEP3 tightly binds to its homologous DNA locus to form a RNA, DNA hybrid, while linear RNA with the same sequence binds weakly to the DNA. Studies have shown that the formation of the circRNA mentioned above, DNA complex, forced the transcription to suspend, resulting in the enhanced splicing efficiency of SEP3 exon-skipped mRNA (42).
Also, ci-ankrd52 belonging to ciRNA participates in prolonging the Pol II mechanism and positively regulates Pol II transcription by accumulating up to its transcription site. The knockout of ci-ankrd52 reduces the expression of its parent gene (3). These studies taken altogether show that some nuclear-localized circRNAs can regulate gene expression at the transcription and splicing levels.
circRNA can translate proteins
Although most circRNAs are ncRNAs, some circRNAs containing internal ribosome entry site elements (IRES) (43-49) or prokaryotic ribosome binding sites (50) are able to encode proteins that differ from their corresponding canonical transcripts. There is a circRNA database named circRNADb containing 32,914 human ecircRNAs, which provides detailed information about circRNA, including genomic sequences, ORFs, and IRES, to predict the translatability of certain circRNAs (51). circZNF609 was found to be translated and function to control myoblast proliferation during muscle differentiation (52). The ribosomal footprint from the fly's head also showed that a group of circRNAs are related to translated ribosomes, and circMbl can produce proteins (6). It is worth noting that IRES is embedded in circMbl and circZNF609 to allow cap-independent translation. Since the ribosome-associated circRNA uses the same start codon as its host mRNA, this circRNA-derived polypeptide may have a similar function to the mRNA-encoded protein or serve as a main negative competitor (52,53).
In addition to IRES, N6-methyladenosine (m6A) modification can also drive circRNA translation. m6A modification has been reported to be the most abundant base modification of RNA and can promote translation of circRNA from reporter genes and endogenous gene loci. circRNA has been identified as being rich in m6A consensus motifs, and a single m6A site is enough to drive translation initiation. The m6A-driven translation is started by eIF4G2 and m6A reader YTHDF3, enhanced by the methyltransferase METTL3/14, inhibited by the demethylase FTO, and upregulated during heat shock (54). Also, m6A-driven translation of circRNA is widespread, and many endogenous circRNAs have translation potential, which indicates the role circRNA-derived proteins potentially hold in cellular stress responses. Cell starvation, for example, leads to enhanced translation of circMbl (53), and the translation of GFP in m6A-rich circRNA plasmids can be promoted by heat shock (54). However, only a small part of circRNA is associated with polysomes, and the cap-independent initiation efficiency is low. Therefore, the products of circRNA translation may be limited (2,34,54,55).
circRNA-derived pseudogenes
Pseudogenes typically originate from the process by which linear mRNAs are integrated into their host genome. Dozens of circRNA-derived pseudogenes have been found in the genomes of humans and mice (56,57). Among them, dozens of pseudogenes derived from circRFWD2 were found in mice, and high-density long terminal repeats (LTR) were found in the flanking regions, indicating that the retrotransposition process of circRFWD2 and LTR are related. Moreover, the insertion of reverse-transcribed circRNA may potentially disrupt the integrity of the host genome. For example, the circSATB1-derived pseudolocus overlaps with CCCTC binding factor (CTCF) and Rad21 binding sites in several mouse cell lines. This CTCF binding is specific in the circSATB1-derived pseudogene region, but is not specific in its original SATB1 region (58).
circRNA as potential biomarkers
As mentioned above, circRNA is abundant and conserved among species. At the same time, it is stably expressed in saliva, blood, exosomes, and shows tissue/developmental stage specificity, which makes it a potential biomarker (58-60). Besides, circRNA is more straightforward to detect than the small number of miRNAs. A variety of circRNAs have been considered as potential biomarkers. In laryngeal squamous cell carcinoma (LSCC), for example, circ_100855 is significantly upregulated, while circ_104912 is significantly downregulated. Their expression is significantly related to tumor stage and cervical lymph node metastasis, suggesting that they could be used as potential new biomarkers in LSCC tumorigenesis (61). Other studies have found that circBRAF can be used as a biomarker to predict the pathological grade and prognosis of glioma patients (62). In bladder cancer, circTCF25 overexpression may promote proliferation and migration and could serve as a novel potential marker (63). Recent studies have shown that mature cancer-related chromosomal translocations produce fusion circRNAs (f-circRNAs), which are produced by transcriptional exons of different genes affected by translocations. When f-circRNA is combined with other carcinogenic stimulants, it can take on a decisive role in promoting leukemia progression (64). f-circM9 and f-circPR are a fusion of the PML/RARa and MLL genes. Knocking out f-circM9 and f-circPR can lead to apoptosis of tumor cells and increase their sensitivity to drugs such as arsenic, which indicates that f-circM9 and f-circPR play a carcinogenic role in hematological malignancies (64). Other studies have found that the circRNA content in human platelets is 17- to 188-fold that of nucleated tissues, mainly because circRNA is more stable than linear mRNA and is not easily degraded (65). Also, circRNA can also be used as ceRNA in the regulation of miRNA on platelets (66). In summary, circRNA can be used as a potential biomarker for the diagnosis, treatment, and prognosis of cancer patients.
circRNA in tumor immunity
Based on the above, circRNAs have many biological functions and play a critical regulatory role in tumorigenesis and development. The immune system plays a dual role in tumor occurrence and development. On the one hand, the immune system can resist tumorigenesis through innate and acquired immunity; on the other hand, tumor cells can escape the recognition and attack by the immune system through the formation of a specialized immune suppressive microenvironment or other mechanisms to generate immune escape. The tumor immune suppressive microenvironment comprises immunosuppressive molecules, matrix components, and immune suppressor cells. These immune suppressor cells include tumor-associated macrophages (TAMs), regulatory T cells (Tregs), and myeloid-derived suppressor cells (MDSCs). An intricate interplay between immune suppressor cells occurs throughout tumor progression. MDSCs, as one example, interfere with the body’s innate immunity by interacting with natural killer (NK) cells and NKT cells; concurrently, they transform macrophages into M2 macrophages with immunosuppressive effects to regulate anti-tumor immunity (67). Given the complex role of the body’s immune system in cancer, and the scarcity of research on tumor immune regulation by circRNA, we systematically summarize the functions of circRNA in tumor immunoregulation under four headings below (Figure 3).
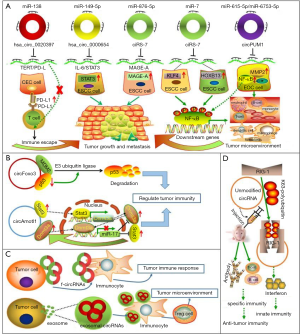
Function as a miRNA sponge to regulate tumor immunity
CircRNA has been reported to regulate tumor immunity via the circRNA-miRNA-mRNA axis (17). For instance, circRNA hsa_circ_0020397 has been revealed to sponge miR-138 and inhibit miR-138 activity, resulting in its downstream target gene telomerase reverse transcriptase (TERT) and programmed death-ligand 1 (PD-L1) enhanced expression in colorectal cancer (CRC) cells. After this, the highly expressed PD-L1 binds to PD-1 in CRC, causing T cell inactivation and tumor immune escape (68). Hsa_circ_0000654 was also identified as an oncogenic circRNA in esophageal squamous cell carcinoma (ESCC) through sponging miR-149-5p and the subsequent indirect activation of the IL-6/STAT3 signaling pathway, which has been widely certificated to be involved in tumor immune processes (69). Moreover, the well-known circRNA, ciRS-7, could accelerate ESCC progression by sponging miR-876-5p to enhance the tumor antigen melanoma-associated antigens-A (MAGE-A) family expression (70).
As is widely understood, the tumor inflammatory microenvironment plays a crucial role in tumor immunity. In the inflammatory microenvironment of tumors, continuous inflammatory reactions result in abnormal activation of the nuclear factor kappa B (NF-κB) pathway and the promotion of tumor development (71). Some studies have shown that certain circRNAs could indirectly affect the NF-κB pathway to regulate tumor immunity. For example, ciRS-7 was found to trigger the migration and invasion of ESCC cells via miR-7/KLF4 and NF-κB signals (72). Moreover, ciRS-7 could also function as a miR-7 sponge and then reactivate the HOXB13-mediated NF-κB pathway, inducing malignant progression of ESCC (11). Also, circRNA-000911 plays an anti-oncogenic role in breast cancer through sponging miR-449a to activate the Notch1 and NF-κB signaling pathways (73). By sponging miR-615-5p and miR-6753-5p, circPUM1 upregulated the expression of NF-κB and MMP2 to exert its carcinogenic effects in epithelial ovarian cancer (EOC) cells (74). We have summarized the function of circRNA as a miRNA sponge to regulate tumor immunity in Table 1.

Full table
Function as RBP sponge to regulate tumor immunity
Unlike the points dicussed above, the regulation of circRNA as an RBP sponge in tumor immunity has rarely been reported. circAmotl1 has been shown to interact with Stat3 and Dnmt3a, which are both the target gene of miR-17-5p. Dnmt3a inhibits miR-17-5p expression through methylation of the miR-17 promoter, and the decreased miR-17-5p induces enhanced Stat3 level in tumor immunity regulation (17,75). Besides, circFoxo3 could bind to MDM2 and p53, resulting in p53 ubiquitination and later degradation, which has been widely reported to regulate tumor immunity and progression (17,76,77). Although there is not enough research on circRNA as an RBP sponge to participate in tumor immunity so far, the above cases suggest that circRNA can regulate tumor immunity by interacting with some immune-related proteins like Stat3 and p53.
Function as tumor antigens to regulate tumor immunity
It was reported that the transfection of exogenous circRNAs into mammalian cells could effectively induce innate immunity and resist viral infection (78). Moreover, another study showed that endogenous circRNAs are prone to form 16-26bp imperfect RNA duplexes and serve as inhibitors of the double-stranded RNA (dsRNA)-activated protein kinases (PKRs) associated with innate immunity (79). During tumorigenesis, cancer-associated chromosomal translocations may produce some novel aberrant fusion circRNAs, that are f-circRNAs, which may be conveyed to immunocytes and then induce tumor immune response (59).
Besides, some exosomal circRNAs may also function as tumor antigens to regulate tumor immunity. For instance, there was a study that revealed that compared with the cellular circRNAs, the expression profile of exosomal circRNAs was more abundant based on RNA-Seq data in three KRAS mutant CRC cell lines (80). Exosomes, as a medium of cell communication, could be easily detected in serum and body fluids, representing a potential tumor biomarker (81). Together with the expression enrichment of circRNAs in exosomes, the exosomal circRNAs play a potential role in the modulation of intercellular connection and tumor microenvironment (17,82). Furthermore, exosomal circRNAs were found to modulate the homeostasis and function of regulatory T (Treg) cells (83).
Function as vaccine adjuvants to induce anti-tumor immunity
Tumor vaccines activate tumor-specific immune responses through tumor-associated antigens to kill and clear tumor cells, which have been widely applied for cancer immunotherapy (84). As a critical part of many tumor vaccines, adjuvants can increase the intensity, breadth, quality, and longevity of the immune response to tumor antigens (85). A recent study demonstrated that unmodified foreign circRNAs could act as potent adjuvants to induce antigen-specific T and B cell activation, antibody production, and anti-tumor immunity. In contrast, m6A modification of the circRNAs recognized as “self” may inhibit innate immunity and adjuvant activity via m6A reader YTHDF2 (86). This study supplies a powerful insight for breakthroughs regarding circRNAs in tumor immunotherapy.
Discussion
This review summarizes the formation mechanism, key features, biological functions, and research advances in tumor immunity of circRNAs. Although circRNAs were initially considered as a by-product of mis-splicing, with the development of high-throughput sequencing in recent years, more and more circRNAs have been discovered and characterized. circRNA can function as a miRNA and RBP sponge, and its inherent closed-loop structure makes it relatively stable in tissue and blood, making it more likely to be a biomarker for tumor diagnosis, treatment, and prognosis. Although most circRNAs are non-coding RNAs, some circRNAs have been found to encode proteins and may perform specific biological functions. Moreover, some circRNAs take part in tumor immunity regulation and may serve as potential molecular targets for tumor immunotherapy.
Currently, tumor immunotherapy plays an increasingly crucial role in cancer treatment. However, non-coding RNA, especially circRNA. is still not commonly applied in tumor immunotherapy. Given the increasingly improved status of circRNA in tumors, the in-depth exploration of the regulatory effect of circRNA on tumor immunity can not only unearth more novel tumor biomarkers and benefits for tumor diagnosis, treatment, and prognosis, but it could also dig out new immunotherapy targets, providing more effective methods for the prevention and treatment of various tumors in the future.
Overall, this review supplies a new perspective on the molecular mechanism and clinical application of circRNAs in the field of tumor immunity. It has specific scientific significance for attracting more scholars to explore the biological potential of circRNAs further.
Acknowledgments
Funding: This work was supported by the National Nature Science Foundation of China (No. 81972722).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-19-4751). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489:101-8. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell 2013;51:792-806. [Crossref] [PubMed]
- Du WW, Zhang C, Yang W, et al. Identifying and Characterizing circRNA-Protein Interaction. Theranostics 2017;7:4183-91. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. [Crossref] [PubMed]
- Costelli P. A circRNA/miR axis contributes to hypoxia-induced cardiomyocyte death. Transl Cancer Res 2018;7:S38-40. [Crossref]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55-66. [Crossref] [PubMed]
- Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res 2013;73:5609-12. [Crossref] [PubMed]
- Cheng X, Qiu J, Wang S, et al. Comprehensive circular RNA profiling identifies CircFAM120A as a new biomarker of hypoxic lung adenocarcinoma. Ann Transl Med 2019;7:442. [Crossref] [PubMed]
- Sang M, Meng L, Liu S, et al. Circular RNA ciRS-7 Maintains Metastatic Phenotypes as a ceRNA of miR-1299 to Target MMPs. Mol Cancer Res 2018;16:1665-75. [Crossref] [PubMed]
- Li RC, Ke S, Meng FK, et al. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis 2018;9:838. [Crossref] [PubMed]
- Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis 2018;9:417. [Crossref] [PubMed]
- Sun X, Wang M, Xu R, et al. Prognostic model based on circular RNA circPDK1 for resected lung squamous cell carcinoma. Transl Lung Cancer Res 2019;8:907-19. [Crossref] [PubMed]
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 2011;331:1565-70. [Crossref] [PubMed]
- Starnes CO. Coley’s toxins in perspective. Nature 1992;357:11-2. [Crossref] [PubMed]
- Cortez MA, Anfossi S, Ramapriyan R, et al. Role of miRNAs in immune responses and immunotherapy in cancer. Genes Chromosomes Cancer 2019;58:244-53. [Crossref] [PubMed]
- Xu Z, Li P, Fan L, et al. The Potential Role of circRNA in Tumor Immunity Regulation and Immunotherapy. Front Immunol 2018;9:9. [Crossref] [PubMed]
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol 2015;12:381. [Crossref] [PubMed]
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777. [Crossref] [PubMed]
- Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science 2013;340:440-1. [Crossref] [PubMed]
- Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015;10:170-7. [Crossref] [PubMed]
- Aktaş T, Avşar Ilık İ, Maticzka D, et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 2017;544:115-9. [Crossref] [PubMed]
- Conn SJ, Pillman KA, Toubia J, et al. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell 2015;160:1125-34. [Crossref] [PubMed]
- Li L, Zheng YC, Kayani MUR, et al. Comprehensive analysis of circRNA expression profiles in humans by RAISE. Int J Oncol 2017;51:1625-38. [Crossref] [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Wang PL, Bao Y, Yee MC, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS One 2014;9:e90859. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett 2015;365:141-8. [Crossref] [PubMed]
- Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2015;58:870-85. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-88. [Crossref] [PubMed]
- Huang R, Zhang Y, Han B, et al. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy 2017;13:1722-41. [Crossref] [PubMed]
- Yu CY, Li TC, Wu YY, et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun 2017;8:1149. [Crossref] [PubMed]
- Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016;7:11215. [Crossref] [PubMed]
- Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014;15:409. [Crossref] [PubMed]
- Calabretta S, Richard S. Emerging Roles of Disordered Sequences in RNA-Binding Proteins. Trends Biochem Sci 2015;40:662-72. [Crossref] [PubMed]
- Kim MY, Hur J, Jeong S. Emerging roles of RNA and RNA binding protein network in cancer cells. BMB Rep 2009;42:125-30. [Crossref] [PubMed]
- Zong FY, Fu X, Wei WJ, et al. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet 2014;10:e1004289. [Crossref] [PubMed]
- Zhao Y, Zhang G, Wei M, et al. The tumor suppressing effects of QKI-5 in prostate cancer: A novel diagnostic and prognostic protein. Cancer Biol Ther 2014;15:108-18. [Crossref] [PubMed]
- Ye Z, Jin H, Qian Q. Argonaute 2: A Novel Rising Star in Cancer Research. J Cancer 2015;6:877. [Crossref] [PubMed]
- Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016;44:2846-58. [Crossref] [PubMed]
- Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 2017;14:361-9. [Crossref] [PubMed]
- Conn VM, Hugouvieux V, Nayak A, et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants 2017;3:17053. [Crossref] [PubMed]
- Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 1995;268:415-7. [Crossref] [PubMed]
- Yang Y, Gao X, Zhang M, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis J Natl Cancer Inst 2018;110:304-15. [published correction appears in J Natl Cancer Inst 2018;110:1147]. [Crossref] [PubMed]
- Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 2018;37:1805-14. [Crossref] [PubMed]
- Zhang M, Zhao K, Xu X, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun 2018;9:4475. [Crossref] [PubMed]
- Zheng X, Chen L, Zhou Y, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer 2019;18:47. [Crossref] [PubMed]
- Liang WC, Wong CW, Liang PP, et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol 2019;20:84. [Crossref] [PubMed]
- Xia X, Li X, Li F, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer 2019;18:131. [Crossref] [PubMed]
- Perriman R, Ares MJ. Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA 1998;4:1047-54. [Crossref] [PubMed]
- Chen X, Han P, Zhou T, et al. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep 2016;6:34985. [Crossref] [PubMed]
- Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 2017;66:22-37.e9. [Crossref] [PubMed]
- Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 2017;27:626-41. [Crossref] [PubMed]
- Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell 2018;71:428-42. [Crossref] [PubMed]
- Zhang Z, Harrison PM, Liu Y, et al. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res 2003;13:2541-58. [Crossref] [PubMed]
- Zhang Z, Carriero N, Gerstein M. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet 2004;20:62-7. [Crossref] [PubMed]
- Dong R, Zhang XO, Zhang Y, et al. CircRNA-derived pseudogenes. Cell Res 2016;26:747-50. [Crossref] [PubMed]
- Bahn JH, Zhang Q, Li F, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem 2015;61:221-30. [Crossref] [PubMed]
- Memczak S, Papavasileiou P, Peters O, et al. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS One 2015;10:e0141214. [Crossref] [PubMed]
- Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015;25:981-4. [Crossref] [PubMed]
- Xuan L, Qu L, Zhou H, et al. Circular RNA: A novel biomarker for progressive laryngeal cancer. Am J Transl Res 2016;8:932-9. [PubMed]
- Zhu J, Ye J, Zhang L, et al. Differential Expression of Circular RNAs in Glioblastoma Multiforme and Its Correlation with Prognosis. Transl Oncol 2017;10:271-9. [Crossref] [PubMed]
- Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep 2016;6:30919. [Crossref] [PubMed]
- Guarnerio J, Bezzi M, Jeong JC, et al. Oncogenic role of fusion-circRNAs derived from cancer associated chromosomal translocations. Cell 2016;165:289-302. [Crossref] [PubMed]
- Alhasan AA, Izuogu OG, Al-Balool HH, et al. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood 2016;127:e1-e11. [Crossref] [PubMed]
- Sunderland N, Skroblin P, Barwari T, et al. MicroRNA Biomarkers and Platelet Reactivity: The Clot Thickens. Circ Res 2017;120:418-35. [Crossref] [PubMed]
- Ostrand-Rosenberg S. Myeloid-derived suppressor cells: More mechanisms for inhibiting antitumor immunity. Cancer Immunol Immunother 2010;59:1593-600. [Crossref] [PubMed]
- Zhang XL, Xu LL. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int 2017;41:1056-64. [Crossref] [PubMed]
- Xu Z, Tie X, Li N, et al. Circular RNA hsa_circ_0000654 promotes esophageal squamous cell carcinoma progression by regulating the miR-149-5p/IL-6/STAT3 pathway. IUBMB Life 2020;72:426-39. [Crossref] [PubMed]
- Sang M, Meng L, Sang Y, et al. Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett 2018;426:37-46. [Crossref] [PubMed]
- Yang L. Mechanisms that drive inflammatory tumor microenvironment, tumor heterogeneity, and metastatic progression. Semin Cancer Biol 2017;47:185-95. [Crossref] [PubMed]
- Huang H, Wei L, Qin T, et al. Circular RNA ciRS-7 triggers the migration and invasion of esophageal squamous cell carcinoma via miR-7/KLF4 and NF-κB signals. Cancer Biol Ther 2019;20:73-80. [Crossref] [PubMed]
- Wang H, Xiao Y, Wu L, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol 2018;52:743-54. [PubMed]
- Guan X, Zong ZH, Liu Y, et al. circPUM1 Promotes Tumorigenesis and Progression of Ovarian Cancer by Sponging miR-615-5p and miR-6753-5p. Mol Ther Nucleic Acids 2019;18:882-92. [Crossref] [PubMed]
- Yang ZG, Awan FM, Du WW, et al. The Circular RNA Interacts with STAT3, Increasing Its Nuclear Translocation and Wound Repair by Modulating Dnmt3a and miR-17 Function. Mol Ther 2017;25:2062-74. [Crossref] [PubMed]
- Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ 2017;24:357-70. [Crossref] [PubMed]
- Huang Y, Yu P, Li W, et al. p53 regulates mesenchymal stem cell-mediated tumor suppression in a tumor microenvironment through immune modulation. Oncogene 2014;33:3830-8. [Crossref] [PubMed]
- Chen YG, Kim MV, Chen X, et al. Sensing Self and Foreign Circular RNAs by Intron Identity. Mol Cell 2017;67:228-238.e5. [Crossref] [PubMed]
- Liu CX, Li X, Nan F, et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 2019;177:865-880.e1. [Crossref] [PubMed]
- Dou Y, Cha DJ, Franklin JL, et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep 2016;6:37982. [Crossref] [PubMed]
- Hou J, Jiang W, Zhu L, et al. Circular RNAs and exosomes in cancer: a mysterious connection. Clin Transl Oncol 2018;20:1109-16. [Crossref] [PubMed]
- Milane L, Singh A, Mattheolabakis G, et al. Exosome mediated communication within the tumor microenvironment. J Control Release 2015;219:278-94. [Crossref] [PubMed]
- Li P, Liu C, Yu Z, et al. New insights into regulatory T cells: exosome- and non-coding RNA-mediated regulation of homeostasis and resident Treg cells. Front Immunol 2016;7:574. [Crossref] [PubMed]
- Schlom J, Gulley JL. Vaccines as an integral component of cancer immunotherapy. JAMA 2018;320:2195-6. [Crossref] [PubMed]
- Bowen WS, Svrivastava AK, Batra L, et al. Current challenges for cancer vaccine adjuvant development. Expert Rev Vaccines 2018;17:207-15. [Crossref] [PubMed]
- Chen YG, Chen R, Ahmad S, et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol Cell 2019;76:96-109.e9. [Crossref] [PubMed]


