FOXP3 promotes colorectal carcinoma liver metastases by evaluating MMP9 expression via regulating S-adenosylmethionine metabolism
Introduction
Colorectal cancer (CRC) tends to be one of the major causes of death from cancer. CRC ranks third among males as the most prevalent cancer, and second among females in the world (1,2). Survival rates for CRC can vary according to a variety of factors, especially the clinical stage. CRC patients’ 5-year survival rate is significantly reduced, followed by an increase in Tumor-Node-Metastasis (TNM) staging levels. For example, the 5-year survival rate of patients with localized cancer (stage I) is about 95%, while the 5-year survival rate is about 13% with distant metastasis (stage IV). In CRC, liver is the most common site for distant metastasis. At diagnosis, 25 percent of CRC patients are confirmed to be present with liver metastases (2). The exact mechanism of the CRC liver metastases, however, is still uncertain. To produce successful therapies, therefore, it is important to elucidate the molecular mechanisms and genetic alterations.
FOXP3 is a member of a group of evolutionarily conserved transcriptional regulators (forkhead box proteins) distinguished by a winged-helix DNA-binding domain (3). FOXP3 plays a critical role in the transfer of immune tolerance and the development of the immunosuppressive tumor microenvironment for master regulating the growth and function of regulatory T-cells, notably the CD4+CD25+ subset derived from the thymus (natural Tregs) (4).
An accumulating amount of data has shown that FOXP3 is disrupted in cancer cells. Research has been carried out into the expression of FOXP3 in a variety of human cancer cells (5-8). The majority of studies have indicated that when FOXP3 is expressed, growth benefit is conferred to cancer cells, which correlates with unfavorable prognosis. However, some other reports have suggested the opposite (9,10).
In CRC, Kim et al. suggested a poor prognosis has been associated with a higher level of FOXP3 in colon cancer cells (11). Sun et al. came to the opposite conclusion, in that FOXP3’s high expression was related to a longer overall and disease-free survival rate. The positive rate of expression was significantly associated with the degree of differentiation, depth of infiltration, metastasis of the lymph nodes and staging of pTNM (12). Nonetheless, contradictory events of FOXP3 expression in CRC and the molecular mechanism of regulation of the FOXP3-mediated gene were not fully characterized.
Metabolism dysregulation is a hallmark in cancer cells, being the totality of reactions that produce energy for supporting the cancer cells alive. The essential amino acid methionine is an emergent feature of the cancer metabolism. Methionine is an essential amino acid containing sulfur which, as part of the methionine process, sequentially undergoes catabolism and recycling. In the methionine cycle methionine is transformed into S-adenosyl-methionine (SAM), the fundamental methyl donor. SAM treatment for hepatocellular carcinoma (HCC) induced with different carcinogens and hepatocarcinogenesis protocols in rats can actively prevent tumors from developing (13).
Furthermore, forced evaluation of SAM in human HCC cells was demonstrated to exhibit a suppressive effect in vivo tumorigenicity in mice (14). These findings show that SAM has a tumor prevention effect. However, the mechanisms of SAM responsible for the effects of cancer metastases still need to be discussed.
Here, by using clinical correlation, conducting cell- and xenograft-based investigations, and analyzing transcriptomes and metabolomics, we explored FOXP3 activity in CRC liver metastases. We showed the effect of FOXP3 on the prognosis of CRC, elevated FOXP3 expression associated with poor survival in CRC. Overexpression of FOXP3 facilitated MMP9 expression through the SAM cycle to modulate CRC liver metastasis. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-3287).
Methods
Patients and variables
We retrospectively searched for fresh frozen tissues of CRC patients with liver metastasis collected from Liaoning Cancer Hospital & Institute. All the patients were diagnosed with primary CRC with synchronous liver metastasis or heterochrony liver metastasis. A precise pathological diagnosis was needed for both primary and metastatic lesions. Simultaneously, data were gathered from the patients’ recorded general clinical and comprehensive pathology information. Written informed consent was obtained from each patient, and the the study protocol received approval from the ethics committee of the Liaoning Cancer Hospital & Institute (Approval No.: 20181225).
TCGA cohort and GEO cohort
RNA-seq data of CRC was downloaded from GSE50760. A total of 54 samples were collected, including CRC tissues, adjacent non-tumorous colorectal tissues, and liver metastasis tissues. The expression profiles of 19,067 coding genes were then measured. The data were preprocessed as follows: the FPKM data was downloaded, and the mean value was chosen to be the initial expression value of the gene for multiple transcripts of the same gene. Pathway enrichment analysis was then carried out through the DAVID (https://david.ncifcrf.gov/) online platform (15). Afterward, weighted co-expression networks were constructed, facilitated by the blockwiseModules function in the WGCNA package (https://labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages/WGCNA/Tutorials/) on the R platform (https://www.r-project.org/). TCGA-COAD and TCGA-READ transcriptome cohort data were available from the cBioPortal for Cancer Genomics website (http://www.cbioportal.org/) (16).
Cell lines and cultures
Human colorectal carcinoma RKO cells (ATCC, Manassas, VA, USA) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, GenDEPOT) supplemented with 10% fetal bovine serum (GenDEPOT) in a CO2 incubator at 5% humidity. Human embryonic kidney epithelial cell line 293T was bought from Invitrogen and cultured in RPMI-1640 medium. Cells were transfected with plasmid DNA using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). Cells in the exponential growth phase were used for the assays.
Immunohistochemical (IHC) staining and evaluation
Immunostaining of FOXP3 was carried out with rabbit monoclonal anti-FOXP3 antibody (1:3,000, Cat. ab214, Abcam, USA). Two experienced pathologists independently assessed the staining of a specific protein in one FFPE slide. Staining intensity was scored as follows: 0: no staining; 1: weak; 2: medium; and 3: strong staining. Staining extent was graded from 0 to 4, according to the coverage percentage of immunoreactive tumor cells (0%, 1–25%, 26–50%, 51–75%, 76–100%). The scores of staining intensity and extent were multiplied, and the total IHC score (on a scale of 0 to 12) was calculated. A score of 0 to 3 signified negative staining and a score of 4 to 12 signified positive staining.
Cell invasion assay
Cell invasion was determined by Transwell assay was conducted to determine the cell invasion. The transfected cells were collected and resuspended in serum-free DMEM medium and cultured on the Matrigel-coated upper surface chamber. FBS medium was put in the lower chamber. A cotton swab was used to remove the remaining cells on the upper membrane surface after 24 h incubation. Next, 4% paraformaldehyde was used to fix the cells adhered to the surface of the lower membrane, and 0.1 per cent crystal violet was used for staining. Then, an optical microscope was used to count the cells.
Quantitative real-time PCR
SYBR® Premix Ex Taq™ (Takara, Dalian, China) on Quantstudio™ 12 k Flex Real-time PCR system (Applied Biosystems, Foster City, CA) was used to detect relevant genes. The primer sequences are included in Table S1.

Full table
Western blot
The anti-N-Cadherin (Cell signaling, 1:2,000), anti-E-Cadherin (Cell Signaling, Danvers, MA, USA, 1:2,000), anti-Vimentin (Abcam, 1:1,000), anti-Snail (Cell signaling, 1:2,000), anti-MMP9 (Cell signaling, 1:2,000), anti-GADPH (Santa Cruz, 1:3,000), anti-c-Myc (Santa Cruz, 1:1,000), and anti-Cyclin D1 (Santa Cruz, 1:1,000), antiFOXP3 (Abcam 1:3,000, Cell signaling 1:2,000) served as the primary antibodies. A 1:3,000–5,000 dilution of the HRP-linked anti-IgG (Santa Cruz) was used as the secondary antibody.
Construction of gene co-expression network between DEGs and DELs
A scale-free co-expression network for the hub DEGs and DELs was constructed with the WGCNA R package. Gene expression similarity matrix was formed by calculating the Pearson correlation coefficient between two genes, which was then transformed into adjacency matrix (a threshold power of β=5), and subsequently into a topological matrix. The extent of the relationship between genes/lncRNAs was determined by topological overlap measure (TOM). The overall level of gene expression within the module was indicated through the calculation of module eigengene (ME) to define the first principal component for a module. The key module can be found by calculating Pearson correlation coefficients between ME in each module and sample traits. Hub genes which met the criteria of having a module membership (MM) value of more than 0.9 were chosen.
SAM detection
The Bridge-It® S-Adenosyl Methionine (SAM) Fluorescence Assay Kit was used to assess the SAMe levels in line with the manufacturer’s protocol. The cells that stably expressed scrambled shRNA or FOXP3-shRNA were used to transiently express empty vector or FOXP3-overexpressing plasmid.
DNA methyltransferase (DNMT) inhibition experiment
DNMT inhibitor, SGI-1027, was employed to inhibit DNMT activity. At 24 or 48 h after transfection, 2 µM of this compound was supplemented in culture medium for 24 h. Protein extraction and total RNA isolation were then conducted.
Statistical analysis
Chi-square test (Fisher’s exact test) was used to analyze categorical variables. Two-sided t-test was used to compare continuous variables, and ANOVA analysis was used for multiple sets of continuous variables. Correlation between continuous variables was summarized using Spearman’s rank test. All statistical analyses were performed using R version 3.5.3. Statistical significance was represented by P<0.05.
Results
Clinicopathological characteristics of the cohorts
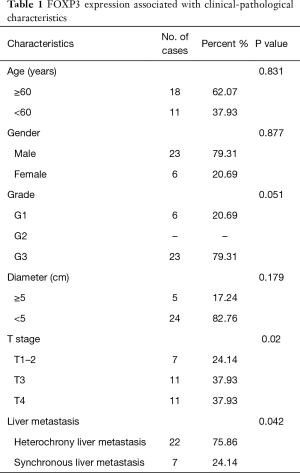
A total of 29 CRC and liver metastasis samples with FOXP3 expression data across patient characteristics were analyzed in this study. Patients with elevated FOXP3 expression in CRC tissue were significantly correlated with T stage (P=0.02) and synchronous liver metastasis (P=0.042) (Table 1). Chi-square test revealed that patients with high FOXP3 expression are prone to progress to a more advanced stage and synchronous liver metastasis than those with low FOXP3 expression.
Full table
FOXP3 is highly expressed in CRC and associated with worse clinical outcome
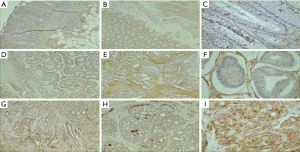
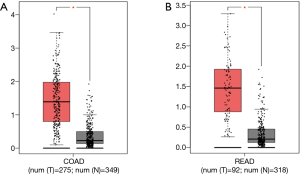
To establish if FOXP3 is expressed in CRC, and to explore if it has any correlation with patient outcome, we carried out immunohistochemistry (IHC) on CRC and colorectal cancer liver metastasis (CRCLM) tissues. FOXP3 immunostaining was seen in CRC and CRCLM cells (Figure 1). FOXP3 expression showed diffuse staining in both the cytoplasm and the nucleus. Statistically, in comparison with normal adjacent tissues, elevated FOXP3 was significantly expressed in cancerous tissues and metastatic liver tissues (P<0.001). DEGs analysis was performed in TCGA-COAD and TCGA-READ to confirm this finding. Figure 2 gives the verification results that FOXP3 is highly expressed in cancer than normal adjacent tissues in the TCGA cohort (P<0.05).
Ectopic expression of FOXP3 facilitates proliferation and migration of CRC cells in vitro and in vivo
To measure the role of ectopic expression of FOXP3 on cancer cells, FOXP3 lentivirus was transfected into CRC cell lines (RKO and HT-29), and Western blot was carried out to validate FOXP3 expression (Figure 3A). We found that overexpression of FOXP3 promotes cell growth in vitro (Figure 3B). Besides, elevated expression of FOXP3 significantly increases cell viability in the invasion and migration of RKO cells (Figure 3C,D,E,F).
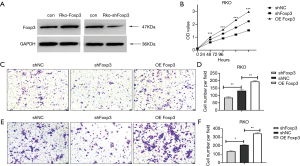
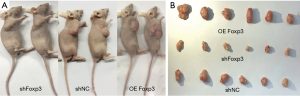
Because cell growth is promoted by FOXP3 in vitro, we next the contribution of FOXP3 in CRC cell growth in vivo. After 18 days of inoculation, the average tumor volume in the overexpression FOXP3-infected group was observed to be 2.6-fold more extensive than that of control group. The tumors of the RKO-FOXP3 and RKO-Control groups of nude mice are shown in Figure 4A and B. Overall, these results illustrated that FOXP3 performs distinct oncogene functions in CRC.
Weighted gene co-expression network analysis of differential genes
Since FOXP3 displays metastasis-related properties, including cell proliferation and migration in CRC, we intended to find a series of combined action genes that promote liver metastasis from a whole transcriptome perspective.
WGCNA was exploited to conduct closely co-expressed high-variant genes (HVGs) into co-expression gene sets. A total of 8,844 HVGs were regarded as the candidate genes and clustered by average linkage hierarchical clustering analysis by transforming adjacency matrix into TOM, and each network module was set with a minimum of 30 genes based on Dynamic Tree Cut standard (Figure 5A,B). Twelve new modules were generated from the calculated eigengenes of each module (Figure 5C). The genes unable to be clustered into other modules were clustered into the gray modules.
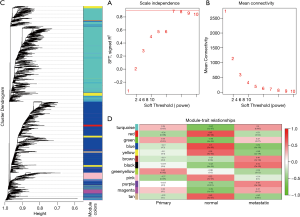
Pearson’s correlation coefficient measured the relationship between each element and the clinical traits (Figure 5D). Genes in yellow, black, brown, magenta, green, blue, and red modules were related to healthy tissues, while genes in green, yellow, purple, pink, and turquoise modules were related to colorectal carcinoma tissues. Meanwhile, the black module was significantly related to the liver metastasis characteristic. Genes included FOXP3 in the black modules are listed in Table S2.

Full table
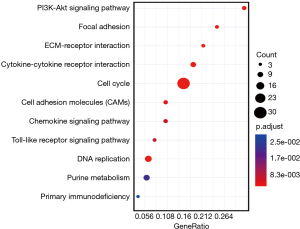
We conducted a KEGG pathway enrichment analysis to reveal the general functional features of the liver metastasis-associated gene set. Notably, pathway enriches in the process of promoting tumorigenicity, including cell cycle, DNA replication, cell adhesion molecules (CAMs), and chemokine signaling pathway (17,18) (Figure 6).
Ectopic expression of FOXP3 activates Wnt/β-catenin pathway in CRC cells
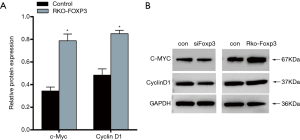
MMP9, Cyclin D1, c-Myc were co-expressed in the black module, which was correlated with CRCLM in transcriptome analysis, and were marker genes in Wnt/β-catenin signaling (19). We further examined the expression of c-Myc and Cyclin D1. Both qPCR (Figure 7A) and Western blot (Figure 7B) revealed the expression of c-Myc and Cylin D1 to be elevated when FOXP3 was ectopically expressed, which indicated that FOXP3 overexpression stimulates the Wnt/β-catenin pathway in CRC cells.
FOXP3 upregulated MMP9 expression through methylthioadenosine (MTA) cycle to modulate colorectal carcinoma metastases
Metastatic colonization formation has been seen to be a significant rate-limiting step during metastatic liver growth (20). This selective pressure may be due in part to the hypoxic microenvironment as well as substantial metabolic activity (21). Therefore, we conducted metabolomics to perform the effect of FOXP3 on tumor metabolism.
Many methionine-related elements of metabolism are linked to cancer. The methionine cycle sees the conversion of methionine to the universal methyl donor SAM, which is converted to S-adenosyl-homocysteine (SAH) after the donation of its methyl group. Through non-target metabolomics analysis, we found that SAM was significantly increased in the siFOXP3 group (Figure 8A). One role methionine plays in controlling cancer-associated phenotypes is mediating gene expression through epigenetic mechanisms. These results suggest that over the SAM cycle, FOXP3 promotes liver metastasis in CRC.
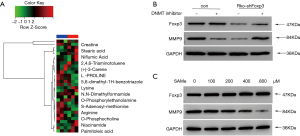
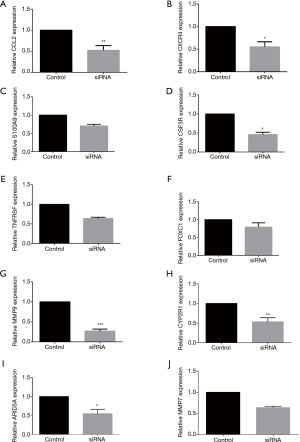
We knocked down FOXP3 via siRNA transfection strategy to shine a light on the role of FOXP3 in the modulation of these co-expression hub genes. The level of mRNA expression of the 10 selected hub genes (Figure S1) was then calculated using qRT-PCR. Results showed that reduction in the expression of FOXP3 significantly reduced the expression of MMP9 more than two-fold (Figure 8B). The matrix metalloproteinases-9, MMP-9, is a significant protease, which is able to regulate ECM remodeling by cleaving many extracellular matrix (ECM) proteins and performs a pivotal task in cancer cell invasion and tumor metastasis. To confirm the impact of FOXP3 on MMP9 expression, western blot analysis, and RT-qPCR were performed in FOXP3 stably expressing cells. The protein and mRNA levels of MMP9 were observed to be markedly increased in cells exhibiting FOXP3 overexpression, as shown in Figure 8C. Conversely, knockdown of FOXP3 significantly reduced MMP9 protein and mRNA expression.
Next, we analyzed whether the downregulation of MMP9 was a downstream event of the SAM cycle according to the results in Figure 8A. As expected, the expression of MMP9 was significantly higher under the supplementation of DNMT inhibitor in cells with FOXP3 overexpression. Also, we analyzed the MMP9 expression level following SAM treatment at various concentrations for 72 h. Accordingly, we noticed a decrease in the MMP9 protein level in a SAM dose-dependent manner. These findings altogether confirmed the hypothesis that FOXP3 negatively regulates SAM and therefore reduced MMP9 promoter methylation to induce its expression.
Discussion
The significant finding from this study is that we indicated FOXP3 acts as a potent oncogene in CRC liver metastasis. Overexpression of FOXP3 enhanced CRC cells with metastatic behavior via activating the Wnt/β-catenin signaling pathway. This resulted in a hyperactivation condition. which is a core characteristic of CRC development. FOXP3-mediated S-adenosylmethionine regulated MMP9 promoter demethylation, upregulating expression of MMP9, which has been widely found to relate to the invasion, metastasis, and angiogenesis pathology of cancers. High FOXP3 expression was significantly associated with poor outcomes in CRC patients. Collectively, our data showed that a high level of FOXP3 expression in cancer cells plays a pivotal role in propelling CRC liver metastasis and decreasing patient survival. It paves a novel way for FOXP3 expression to be able to influence the pathogenesis of CRC liver metastasis via interference with S-adenosylmethionine metabolism.
It is well known that FOXP3 is a specific marker of regulatory T cells (Tregs), and that performs a vital task in the differentiation of Tregs to mediate tumor immune escape (17,22). However, recently its expression has been shown in distinct types of human tumor cells. FOXP3 exhibited paradoxical roles in different types of tumors. Li et al. demonstrated that FOXP3 suppresses angiogenesis of breast cancer by downregulating the expression of VEGF (18). Guo et al. revealed that the expression of FOXP3 was correlated with the grade of gastric lesion, indicating that FOXP3 may have an essential function in gastric cancer formation, development and prognosis (23). Shi et al. suggested that FOXP3 has a suppressive effect on the progression of HCC tumors via the transforming growth factor-beta (TGF-β)/Smad2/3 signaling pathway, which presents FOXP3 as a factor in the prognosis of and a novel target for the optimal treatment of HCC (24).
On the contrary, Tang et al. showed that there is a positive correlation between FOXP3 and vascular endothelial growth factor-C (VEGF-C) expression and lymphangiogenesis in cervical cancer (25). Mechanistically, Gao et al. found that nuclear Galectin-1 (Gal-1) interferes with the binding process between FOXP3 and DNA through its interaction with FOXP3’s FKH domain, which may the possible mechanism behind the loss of the tumor-suppressive properties of FOXP3 in wild-type FOXP3-positive breast cancer (26). In this study, data analysis of our cohort and CRC cells supported our hypothesis; the high expression of FOXP3 detected in tumor tissues from CRC patients led to the activation of the Wnt signaling pathway and decreased overall survival.
KEGG analysis illustrated that prostate cancer samples were enriched with meiotic recombination 11 (MRE11), which is involved in the most significant pathways, including the mitotic spindle, ultraviolet response, and TGF-β signaling pathways. Of the Wnt signaling pathways, the established Wnt/β-catenin pathway has been investigated most extensively. In the established pathway, when there are adequate levels of β-catenin, a fraction of cytoplasmic β-catenin migrates to the nucleus to interact with the LEF-1/T-cell factor (TCF) family transcription factors. As a consequence, these transcription factors stimulate the activation of oncogenes involved in EMT, survival, angiogenesis motility, and invasion, including c-Myc, VEGF, monocyte chemo-attractant protein 1 (CCL2), snail, slug, vimentin, and metalloproteinases (27-30). To this end, FOXP3 was chosen for further study of its potent role in driving metastasis, considering its recognized role in promoting CRC cell migration and invasion.
The relationship between FOXP3 and CRC liver metastasis has seldom been reported, although notably, MRE11 has been confirmed to support a reduced overall survival rate and is believed to be highly expressed in tissues from CRC tumors and liver metastasis.
Metabolomics analysis of CRC cells revealed that the metastasis promotion effect by FOXP3 was mediated through the SAM cycle. Following our results, Luo et al. reported that SAM could effectively exhibit an inhibitive effect on tumor cell growth via the reversal of DNA hypomethylation on oncogene promoters, leading to the downregulation of their expression. As it has no effect on the expression of P16 and other tumor suppressor genes, SAM could potentially be applied in cancer therapy (31). This was suggested in the study of Luo et al. (31). Previous studies also found that SAM acts as a regulator of apoptosis and autophagy in MCF-7 breast cancer cells via the microRNA-specific modulation (32). On the other hand, Shi et al. reveal that SAM presents as evidence of tumor suppression, especially on primary osteosarcoma, but it lacks positive effects on metastatic osteosarcoma (33). All these studies prove that the SAM cycle plays a significant role in tumor growth and development. Further, Greenberg et al. demonstrate that plasma levels of SAM are significantly elevated in lung cancer patients and, in combination with chest CT, may serve as a helpful tool for the early diagnosis of lung cancer (34).
In our study, FOXP3 inhibition was shown to suppress the expression of MMP9, which was characteristic of a downstream event of the SAM cycle. It has been reported that MMP9 can be an essential driver of tumor growth and metastasis in many types of tumors. Bing Xia et al. revealed circular RNA derived from MMP9 facilitates oral squamous cell carcinoma metastasis through regulation of MMP9 mRNA stability (35). Similarly, Diosmetin was found to influence the inhibition of SK-HEP-1 and MHcc97H cell metastasis through the downregulation of MMP-2/9 expression via the PKC/MAPK/MMP pathways (36). Moreover, Chen et al. reported that G-protein-coupled receptor kinase-interacting protein 1 can facilitate tumor progression via the activation of ERK/MMP9 signaling in hepatocellular carcinoma (37). We recognized that FOXP3 affects MMP9 expression is in a SAM-dependent way. SAM can effectively inhibit the tumor cell growth via the reversal of the DNA hypomethylation on oncogene promoters, without influencing the expression of the tumor suppressor genes. These results suggest that SAM could potentially be an option for cancer treatment.
At the same time, our study has limitations as follows. Firstly, this study is a retrospective small sample study. Besides, the revelation of detailed mechanism between FOXP3 and metastasis of CRC as well as specific relation of SAM and MMP9 need further experiments to confirm.
In conclusion, our study reveals the relationship that increased FOXP3 expression has with liver metastasis and poor survival in CRC patients. These data indicate that FOXP3 may function as an oncogene and that it holds promise as a biomarker for CRC patients with liver metastasis. More validation cohorts and further elucidation are needed so that all of the values of FOXP3 in cancer cells and its clinical application for CRC liver metastasis can be established.
Acknowledgments
Funding: Science and Technology Planning Project of Shenyang (No. 191124088); Science and Technology Planning Project of Liaoning Province of China (No. 201800449); Natural Science Foundation of Liaoning Province (No. 20180550781). Scientific research foundation for the introduction of talents, Liaoning Cancer Hospital & Institute (No. Z1702).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-3287
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3287
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3287). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Written informed consent was obtained from each patient, and the study protocol received approval from the Ethics Committee of the Liaoning Cancer Hospital & Institute (Approval No.: 20181225).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Henry JT, Johnson B. Current and evolving biomarkers for precision oncology in the management of metastatic colorectal cancer. Chin Clin Oncol 2019;8:49. [Crossref] [PubMed]
- Chen X, Yan S, Zhao H, et al. The safety and feasibility of a single incision in simultaneous resection for patients with colorectal cancer liver metastases. Ann Transl Med 2019;7:547. [Crossref] [PubMed]
- Lal G, Bromberg JS. Epigenetic mechanisms of regulation of Foxp3 expression. Blood 2009;114:3727-35. [Crossref] [PubMed]
- Hinz S, Pagerols-Raluy L, Oberg HH, et al. Foxp3 expression in pancreatic carcinoma cells as a novel mechanism of immune evasion in cancer. Cancer Res 2007;67:8344-50. [Crossref] [PubMed]
- Wang L, Liu R, Li W, et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell 2009;16:336-46. [Crossref] [PubMed]
- Weller P, Bankfalvi A, Gu X, et al. The role of tumour FoxP3 as prognostic marker in different subtypes of head and neck cancer. Eur J Cancer 2014;50:1291-300. [Crossref] [PubMed]
- Zhang HY, Sun H. Up-regulation of Foxp3 inhibits cell proliferation, migration and invasion in epithelial ovarian cancer. Cancer Lett 2010;287:91-7. [Crossref] [PubMed]
- Ma GF, Miao Q, Liu YM, et al. High FoxP3 expression in tumour cells predicts better survival in gastric cancer and its role in tumour microenvironment. Br J Cancer 2014;110:1552-60. [Crossref] [PubMed]
- Suh JH, Won KY, Kim GY, et al. Expression of tumoral FOXP3 in gastric adenocarcinoma is associated with favorable clinicopathological variables and related with Hippo pathway. Int J Clin Exp Pathol 2015;8:14608-18. [PubMed]
- Kim M, Grimmig T, Grimm M, et al. Expression of Foxp3 in colorectal cancer but not in Treg cells correlates with disease progression in patients with colorectal cancer. PLoS One 2013;8:e53630. [Crossref] [PubMed]
- Sun X, Feng Z, Wang Y, et al. Expression of Foxp3 and its prognostic significance in colorectal cancer. Int J Immunopathol Pharmacol 2017;30:201-6. [Crossref] [PubMed]
- Pascale RM, Simile MM, De Miglio MR, et al. Chemoprevention of hepatocarcino-genesis: S-adenosyl-L-methionine. Alcohol 2002;27:193-8. [Crossref] [PubMed]
- Li J, Ramani K, Sun Z, et al. Forced expression of methionine adenosyltransferase 1A in human hepatoma cells suppresses in vivo tumorigenicity in mice. Am J Pathol 2010;176:2456-66. [Crossref] [PubMed]
- Skrlj B, Kunej T. Computational identification of non-synonymous polymorphisms within regions corresponding to protein interaction sites. Comput Biol Med 2016;79:30-35. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Plitas G, Rudensky AY, Regulatory T. Cells: Differentiation and Function. Cancer Immunol Res 2016;4:721-5. [Crossref] [PubMed]
- Li X, Gao Y, Li J, et al. FOXP3 inhibits angiogenesis by downregulating VEGF in breast cancer. Cell Death Dis 2018;9:744. [Crossref] [PubMed]
- Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of be-ta-catenin-independent Wnt signaling. Dev Cell 2003;5:367-77. [Crossref] [PubMed]
- Kauffman EC, Robinson VL, Stadler WM, et al. Metastasis sup-pression: the evolving role of metastasis suppressor genes for regulating cancer cell growth at the secondary site. J Urol 2003;169:1122-33. [Crossref] [PubMed]
- Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic in-stability. Nat Rev Cancer 2008;8:180-92. [Crossref] [PubMed]
- Mohr A, Atif M, Balderas R, et al. The role of FOXP3+ regulatory T cells in human autoimmune and inflammatory diseases. Clin Exp Immunol 2019;197:24-35. [PubMed]
- Guo G, He Z, Shi Z. Correlation between FOXP3 expression and gastric cancer. Oncol Lett 2016;12:1554-8. [Crossref] [PubMed]
- Shi JY, Ma LJ, Zhang JW, et al. FOXP3 Is a HCC suppressor gene and Acts through regulating the TGF-β/Smad2/3 signaling pathway. BMC Cancer 2017;17:648. [Crossref] [PubMed]
- Tang J, Yang Z, Wang Z, et al. Foxp3 is correlated with VEGF-C expression and lymphangiogenesis in cervical cancer. World J Surg Oncol 2017;15:173. [Crossref] [PubMed]
- Gao Y, Li X, Shu Z, et al. Nuclear galectin-1-FOXP3 interaction dampens the tumor-suppressive properties of FOXP3 in breast cancer. Cell Death Dis 2018;9:416. [Crossref] [PubMed]
- Clevers H. Wnt/β-catenin signaling in development and disease. Cell 2006;127:469-80. [Crossref] [PubMed]
- Wang N, Wang ZY, Wang Y, et al. Dietary compound isoliquiritigenin prevents mammary carcinogenesis by inhibiting breast cancer stem cells through WIF1 de-methylation. Oncotarget 2015;6:9854-76. [PubMed]
- Kwon OJ, Valdez JM, Zhang L, et al. Increased Notch signal-ling inhibits anoikis and stimulates proliferation of prostate luminal epithelial cells. Nat Commun 2014;5:4416. [Crossref] [PubMed]
- Przybylo JA, Radisky DC. Matrix metalloproteinase-induced epithelial-mesenchymal transition: tumor progression at Snail’s pace. Int J Biochem Cell Biol 2007;39:1082-8. [Crossref] [PubMed]
- Luo J, Li YN, Wang F, et al. S-adenosylmethionine inhibits the growth of cancer cells by reversing the hypomethylation status of c-myc and H-ras in human gastric cancer and colon cancer. Int J Biol Sci 2010;6:784-95. [Crossref] [PubMed]
- Ilisso CP, Delle Cave D, Mosca L, et al. S-Adenosylmethionine regulates apoptosis and autophagy in MCF-7 breast cancer cells through the modulation of specific microRNAs. Cancer Cell Int 2018;18:197. [Crossref] [PubMed]
- Shi H, Mu WD, Zhang B, et al. Potential role of S-adenosylmethionine in osteosarcoma development. Onco Targets Ther 2016;9:3653-9. [PubMed]
- Greenberg AK, Rimal B, Felner K, et al. S-adenosylmethionine as a biomarker for the early detection of lung cancer. Chest 2007;132:1247-52. [Crossref] [PubMed]
- Xia B, Hong T, He X. A circular RNA derived from MMP9 facilitates oral squamous cell carcinoma metastasis through regulation of MMP9 mRNA stability. Cell Transplant 2019;28:1614-23. [Crossref] [PubMed]
- Liu J, Wen X, Liu B, et al. Diosmetin inhibits the metastasis of hepatocellular carcinoma cells by downregulating the expression levels of MMP-2 and MMP-9. Mol Med Rep 2016;13:2401-8. [Crossref] [PubMed]
- Chen J, Yang P, Yang J, et al. GIT1 is a novel prognostic biomarker and facilitates tumor progression via activating ERK/MMP9 signaling in hepatocellular carcinoma. Onco Targets Ther 2015;8:3731-42. [PubMed]