Analyzing the prognostic value of DKK1 expression in human cancers based on bioinformatics
Introduction
The incidence of cancer is growing rapidly worldwide, and is the one of the major causes of global morbidity and mortality, posing significant challenges to public health (1). Each year in the United States, 1.5 million people are diagnosed with malignancies, and 0.6 million individuals die of cancer (2). Regardless of whether cancer is fatal to patients, it is still an overwhelming condition for many people (3). Gender is one axis along which way cancer incidence can be differentiated. Among males, lung cancer is the most common cancer and the leading cause of cancer death, while for females, breast cancer is the most deadly (4). Despite the rapid development of diagnostic and therapeutic methods that have been introduced into clinical practice, there are still many cancer patients who cannot be cured. It is thus crucial that we gain further insight into the mechanisms of carcinogenesis, and find the potential biomarkers for early diagnosis, accurate prognosis, and targets for therapy in cancers.
We used public databases GEPIA and Oncomine to find the DKK1 differentially express in multiple tumors and has a deep impact on the prognosis of head and neck squamous cell carcinoma (HNSC), LUSC, and pancreatic adenocarcinoma (PAAD). Overexpression of DKK1 indicated adverse OS in bladder urothelial carcinoma (BLCA). The Dickkopf1 (DKK1) gene, a Wnt signaling pathway inhibitor, encodes a protein that belongs to one of the members of the Dickkopf family, and has an important role in several biological contexts and pathways. The Wnt signaling pathway controls tissue development and homeostasis by regulating endogenous stem cells. In recent decades, the theory of cancer stem cells (CSCs) has emerged and yielded a number of impressive achievements in this field. CSCs have been observed in multiple types of cancer including breast cancer (5), pancreatic cancer (6), and acute myeloid leukemia (AML) (7). CSCs are responsible for the resistance of chemotherapy and radiotherapy, along with cancer relapse and metastasis (8,9). Wnt/β-catenin pathway governs the progression of CSCs (10). Thus, aberrant Wnt signaling may play a key role in tumorigenesis and the process of many cancers via affecting the cancer stem cells. DKK1 is the regulator of the Wnt signaling pathway, and many studies have reported on the abnormal expression of DKK1 in multiple cancers (11-13). The function of DKK1 varies across different cancers types. In myeloma, the overexpression DKK1 may be related to osteolytic bone (14), while high DKK1 expression maybe the mechanism behind inducing crizotinib resistance in non-small cell lung cancer (NSCLC) (15). Aufderklamm et al. found that DKK1 could downregulate osteoblast activity to promote the progression of metastatic prostate cancer (16). Interestingly, some researchers found the elevated expression of DKK1 could improve head and neck cancer’s cell sensitivity to cisplatin (17). Collectively, the relevant research suggests that DKK1 is a potential biomarker for the prognosis or diagnosis of malignancies. However, with DKK1 acting in a protective or detrimental role according to the cancer type, certainty concerning its function in specific cancers remains elusive. Therefore, a classification of the relation between DKK1 expression with cancer patients‘ prognosis and tumorigenesis is necessary.
In this study, we comprehensively assessed DKK1 expression and its value in predicting cancer patients’ prognosis via use of the Oncomine, Gene Expression Profiling Interactive Analysis (GEPIA), UALCAN, and DriverDBv3 public databases. In order to investigate the potential biological functions and pathways of DKK1 in cancers, we also used GeneMANIA and the Metascape interactive online website to analyze the functional network of DKK1. Results of these analyses indicated that DKK1 may be a valuable biomarker for predicting prognosis in head and neck squamous cell carcinoma (HNSC), lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), and bladder urothelial carcinoma (BLCA). There is further potential for DKK1 to be used as a diagnostic marker or therapeutic target for these identified cancer types.
Methods
Oncomine database analysis
The Oncomine database is the largest oncogene chip database and integrated data mining tool, containing 715 gene expression datasets from 86,733 cancers and normal samples (18). The mRNA expression differences of the DKK1 gene between tumors and normal tissues in distinct types of cancer were determined using the Oncomine database. The results from this analysis are displayed with the P value of 0.01, fold change of 1.5, and all gene rankings.
Gene expression profiling interactive analysis (GEPIA) analysis
GEPIA is a commonly used interactive website that plots expression profiles of given genes. GEPIA, which contains 9,736 tumors and 8,587 normal tissues from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) database, can perform survival analysis based oncogene expression levels according to user-defined sample elections and methods (19). We used GEPIA to determine the expression level of the DKK1 gene in different cancer types. The correlation between DKK1 expression and prognosis of overall survival (OS) and disease-free survival (DFS) in different cancers was also analyzed by GEPIA. The hazard ratio (HR) and P or Cox P values from a log-rank test were included in the plot.
UALCAN database analysis
UALCAN is web resource useful for analyzing cancer data. UALCAN provides graphs and plots depicting gene expression and survival curves, evaluates promoter DNA methylation information, and conducts pan-cancer gene expression analysis (20). In the present study, we used UALCAN to find the patient survival information across different types of cancer based on DKK1 gene expression.
DriverDBv3 database analysis
DriverDBv3 is a cancer omics database that incorporates RNA expression, microRNA (miRNA) expression, methylation, copy number variation, and somatic mutation (21). One of the useful features is that this database allows customizable cancer and gene analysis enabling researchers to analyze the correlations between cancers and driver genes. DriverDBv3 is also able to find genes and present them with different molecular features via published bioinformatics algorithms. We used DriverDBv3 to examine the prognostic potential of DKK1gene expression levels in different human cancers. Survival-relevant expressions, with a log-rank P value <0.05 were considered significant.
GeneMANIA analysis
GeneMANIA is a commonly used website for performing protein-protein interaction (PPI) network analysis and predicting the function of preferred genes (22). This user-friendly online tool can display gene or gene lists using bioinformatics methods, including gene co-expression, physical interaction, gene co-location, gene enrichment analysis, and website prediction. We predicted the function of the DKK1 gene and visualized the gene networks through GeneMANIA.
Functional enrichment analysis via Metascape and Cytoscape
The interactive genes of DKK1, as constructed by the GeneMANIA, were all input into Metascape and Cytoscape for further gene annotation and analysis (23). Cytoscape was used to visualize the results of the protein interaction network (24). Metascape was applied to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, and mCODE, a Cytoscape plugin, was used to obtain the core modules of the genes associated with DKK1. GO term analysis includes biological processes (BP), cellular components (CCs), and molecular function (MF). P values <0.05 were considered as statistically significant.
Results
DKK1 expression levels in different types of human cancers
To find differences in DKK1 expression in tumor and normal tissues, the DKK1 gene expression profiles across tumor samples and paired normal tissues were determined using the Oncomine database. This database contained a total of 400 unique analyses for DKK1. Up-expression of
DKK1 was observed in cancers based on 6 significant unique analyses, while down-expression was observed in 3 analyses. The DKK1expression was higher in the samples of brain and central nervous system cancer, head and neck cancer, liver cancer, and pancreatic cancer than in the samples of normal tissues. In addition, down-expression was found in bladder cancer and prostate cancer (Figure 1A). These analyses suggest that DKK1 has an elevated expression in most cancers. To further evaluate the expression level of DKK1 between tumors and normal tissues in various cancers, we used the GEPIA website, whose data is based on abundant samples from TCGA and GTEx databases, to analyze the data of RNA sequencing expression. The differential expression level of DKK1 is shown in Figure 1B. Compared with normal samples, a higher DKK1 expression was seen in ESCA, HNSC, LUSC, and PAAD, but a lower expression was seen in bladder urothelial carcinoma (BLCA) and cutaneous melanoma .
Prognostic potential of DKK1 in different cancers
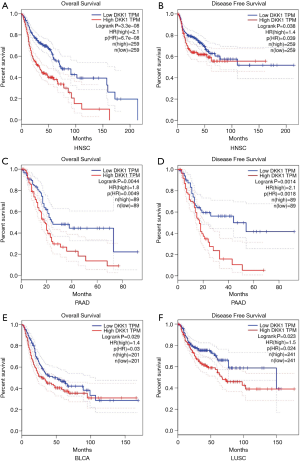
To explore whether an association between DKK1 expression level and prognosis in cancer patients exists, we used GEPIA to analyze the prognostic value of DKK1 in cancers within the RNA sequencing data in TCGA. The results showed that DKK1 overexpression was associated with poorer prognosis of OS and DFS in HNSC (OS log-rank P=3.3e−08, HR =2.1; DFS log-rank P=0.038, HR =1.4) and PAAD (OS log-rank P=0.0044, HR =1.8; DFS log-rank P=0.0014, HR =2.1), and the OS but not DFS of LUSC (DFS log-rank P=0.023, HR =1.5) and BLCA (OS log-rank P=0.029, HR =1.4) (Figure 2). DKK1 expression levels showed weak associations with ESCA, skin cutaneous melanoma (SKM), liver cancer, bladder cancer, prostate cancer, and brain and central nervous system cancers. These results imply that the overexpression of DKK1 can lead to adverse prognosis in HNSC, PAAD, LUSC, and BLCA.
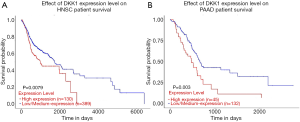
The UALCAN database was also used to assess the prognostic values of DKK1 and further define the cancer types. The association between DKK1 expression levels and significant prognosis information in determining cancers is shown in Figure 3. Notably, DKK1 expression levels jada considerable effect on the prognosis in HNSC and PADD. Results showed that higher DKK1 expression levels indicated poor outcomes of HNSC and PAAD patients. In order to evaluate the role of DKK1 expression levels that impacted the prognosis of different cancers, we also used the DriverDBv3 database to create a survival curve for our selected cancer types. Overexpression of DKK1 was related to poor prognosis in PAAD [disease free interval (DFI) log-rank P=0.0044, HR =3.11; platinum-free treatment interval (PFI) log-rank P=0.0297, HR =1.58; OS log-rank P=0.00453, HR =1.85, log-rank P=0.00251, HR =1.91], OS in HNSC (OS log-rank P=0.0205, HR =1.41), and shortened PFI time in LUSC (log-rank P=0.0161, HR =1.54). However, the survival rate of BLCA patients was not found to be significantly associated with DKK1 expression. Based on the 3 database analyses, the results indicated that DKK1 expression levels can be a potential biomarker for predicting prognosis in PAAD and HNSC patients. DKK1 expression levels could also be a biomarker for deciding whether to take positive action in LUSC therapy (Figure 4).
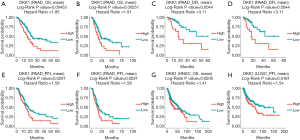
Re-analysis of the prognostic value of DKK1 expression for cancer
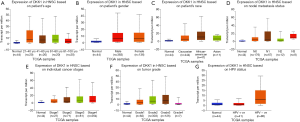
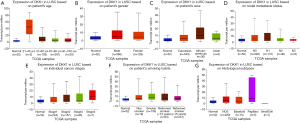
In order to further assess DKK1 expression levels for cancers in which DKK1 expression showed potential prognostic value, we used the UALCAN database to perform subgroup analyses of different clinical features of defining cancers including those of age, gender, cancer stage, race, life habit, etc. Further analyses showed that the transcription levels of DKK1 were significantly higher in HNSC, PAAD, and LUSC than in normal samples. Subgroup analyses showed that DKK1 expression levels were higher in features including age from 20 to 40 years, African-American race, and grade 3, than in other clinical features (Figures 5-7). These findings suggest that DKK1 expression levels can serve as a potential diagnostic biomarker in certain cancer types.

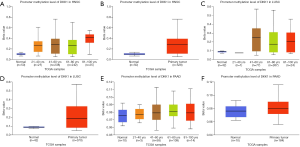
Prognostic value of promoter methylation levels of DKK1
We next used the UALCAN database to explore the level of DKK1 promoter methylation for the specified cancers and investigated the association of promoter DNA methylation with DKK1 expression levels. The results showed that the promoter methylation levels of DKK1 in all the defined cancers were higher than in the normal samples. To explore the factors that affect the levels of promoter methylation DKK1, we analyzed subgroup promoter DNA methylation of DKK1 according to different clinical features. The subgroup analysis results showed the promoter methylation of DKK1 was possibly impacted by age in the specified cancers (Figure 8). These results indicate that lower promoter DNA methylation can lead to the up-expression of DKK1 in HNSC, LUSC, and PAAD.
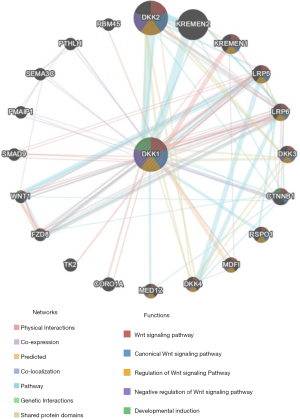
PPI network analysis via GeneMANIA
The PPI network analysis revealed an interaction among genes for DKK1. The functions of genes enriched for DKK1 were responsible for the Wnt signaling pathway, canonical Wnt signaling pathway, regulation of Wnt signaling pathway, negative regulation of Wnt signaling pathway, and developmental induction (Figure 9). The results of the PPI network analysis indicated that the function of DKK1 is s a modulator of the WNT signaling pathway.
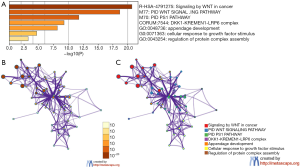
Functional enrichment analyses of DKK
To further predict the enrichment function information of the interactive genes of DKK1, we performed analyses of GO and KEGG pathway in Metascape. This showed that DKK1 interactive genes participate in multiple BP, CCs, and MFs. We found that DKK1-KREMEN1-LRP6 complex, regulation of protein complex assembly, cellular response to growth factor stimulus, and appendage development were heavily regulated by DKK1 and its interactive genes (Figure 10A). The significantly enriched KEGG pathways included signaling by Wnt in cancer, PID PS1 pathway, and PID Wnt signaling pathway. These results demonstrate that DKK1 acts prominently in protein complex assembly, cellular response to growth factor stimulus, regulating the WNT signaling pathway in cancers, and other essential biological processes. To determine the relations within the enriched terms, we selected a subset of terms and established a network plot. Edges linked the terms, including a similarity >0.3. Then, we visualized the network using Cytoscape, as shown in Figure 10B,C. Each node represents an enriched term and is colored by the cluster-ID. The enriched terms included DKK1-KREMEN1-LRP6 complex, regulation of protein complex assembly, cellular response to growth factor stimulus, appendage development, signaling by Wnt in cancer, PID PS1 pathway, and PID Wnt signaling pathway.
Analyzing aberrant DKK1 types of subgroups lung cancer in the cBioPortal
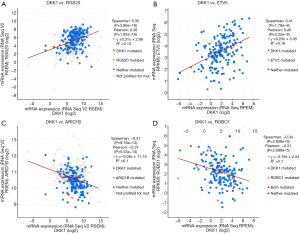
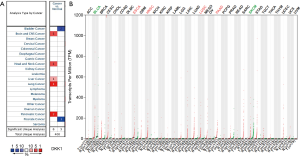
The cBioPortal for Cancer Genomics (http://cbioportal.org) provides a Web resource for exploring, visualizing, and analyzing multidimensional cancer genomics data (25). We not only analyzed the aberrant DKK1 expressions in LUSC but also those in lung adenocarcinoma (Figure 11A). We observed that missense is the most aberrant type in lung cancer (Figure 11B). We also found that DKK1 is significantly associated with RG S20, ETV5, ARID1B, and ROBO1 (Figure 12).

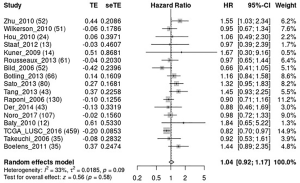
The OS meta-analysis of DKK1 expression in LUSC via the Lung Cancer Explorer (LCE)
To further confirm whether DKK1 can be used as a valuable biomarker for predicting the prognosis LUSC, we performed a meta-analysis of OS for DKK1 expression in LUSC via LCE (Figure 13) (26). The results of the meta-analysis showed that DKK1 expression is marginally related to the OS of LUSC.
Discussion
Despite the improvements in diagnosis and therapy, cancer remains one of the most threatening human diseases. The understanding of the mechanisms of tumorigenesis, and the resistance to chemotherapy, radiotherapy, and immunotherapy are still not sufficiently clear, leading to many cancer patients having short survival times. Therefore, more sensitive, specific, and efficient biomarkers for the diagnosis and therapy of cancers are urgently needed. Thus far, a number of signaling pathways have been identified as playing key roles in cancer-related processes, and among these is the Wnt signaling pathway (27). The fundamental biological functions of the Wnt signaling pathway are cellular maintenance and development. Further, the Wnt signaling pathway regulates normal cell cycle progression, differentiation, migration, with aberrant WNT signaling pathway transduction being found in many cancer types (28-31). Wnt/beta-catenin signaling has been observed to contribute to the promotion of multiple cancers. DKK1 gene encodes protein, and is the most notable member of the Dickkopf family (DKK1–DKK4) as it inhibits Wnt/beta-catenin signaling transduction. Indeed, abnormal expression levels of DKK1 have also been observed in a number of cancers.
Based on this premise, we investigated the relation of DKK1 expression with prognosis in certain cancers using bioinformatics. The results showed DKK1 up-expression levels in multiple cancers. The higher expression of DKK1 may be a potential prognostic biomarker in some cancers such as HNSC, PAAD, and LUSC. A higher DKK1 expression has been observed in BLCA compared to normal samples, and the higher levels of DKK1 can increase the risk of regional lymph node and distant metastasis and indicate poor outcome. Meanwhile overexpression of DKK1 may lead to improved NSCLC cell invasion and migration ability in vitro. Furthermore, higher DKK1 expression may be the potential mechanism of cisplatin resistance. Salim et al.’s study showed that DKK1 expression levels in cisplatin-refractory clones were significantly higher than those in the untreated clones. They also found that knockdown of DKK1 could increase ovarian cancer cell sensitivity to cisplatin (32). In our present study, the DKK1 was found to be overexpressed in LUSC (a type of NSCLC), but the expression levels did not show a critical effect on OS in the GEPIA, UALCAN, and DriverDBv3 databases. Unlike OS, the PFS of LUSC patients was significantly different between LUSC samples and healthy samples. Interestingly, another study found that the up-expression of DKK1 could decrease the risk of regional lymph node metastasis for oral cancers, while its down-expression could accelerate oral cancer cell migration and invasiveness (33). In contrast with oral cancer, PAAD cells with higher DKK1 expression were shown to exhibit improved invasion and growth ability in vitro (34). This finding indicates that overexpression of DKK1 may be an adverse biomarker for the outcome of PAAD patients. Other research supports DKK1 as a potential biomarker for PAAD. For instance, one study proved that DKK1 was more sensitive than CA199 for diagnosing pancreatic cancer, and that higher expression levels of DKK1 indicated poor survival (35). Cytoskeleton-associated protein 4 (CKAP4), a DKK1 receptor, is a potential molecular therapeutic target in pancreatic cancer (36). Moreover, the elevated expression of DKK1 is an independent detrimental factor for HNSC (37). These results are nearly consistent with our present study, which found the overexpression of DKK1 in HNSC, PAAD, and LUSC, with higher DKK1 expression being linked with adverse outcomes in HNSC, PAAD, and BLCA patients. Besides these defined cancers, DKK1 overexpression was observed in melanoma (38), and ovarian (39), colon (40), and breast cancer (41). Furthermore, research of an anti-DKK1 monoclonal antibody for multiple myeloma has been performed in vitro, yielding promising results (42). We not only analyzed the expression of DKK1 but also investigated whether different clinical features affect expression levels. The subgroup analysis indicated that different clinical features of DKK1 may have different expression levels. Low promoter methylation, African-American race, and grade 3 cancers subgroups showed a higher DKK1 expression for all defining cancer types. Eating habits, cancer burden, and virus infection also impacted the DKK1 expression. Based on these features, we should carefully identify different conditions and combine DKK1 with other markers and methods to improve diagnostic effectiveness.
In our study, we found the expression of DKK1 to be significantly related to solid tumors in HNSC, LUSC, and PADD. For solid tumors, the important factor that accelerates cancer growth and metastasis to the distant organ is angiogenesis. This process often involves cancer cells losing the epithelial features and acquiring the mesenchymal traits. Angiogenesis and epithelial-to-mesenchymal transition (EMT) are regulated by Wnt signal transduction. Macrophages and another myeloid cells have been shown to provide the ligand for Wnt in angiogenesis and the EMT process (43). The non-canonical Wnt signal pathway not only regulates EMT but also improves the production of proteolytic enzyme secretion, like matrix metallopeptidase, enabling local invasion and promoting distant metastasis (44,45). Furthermore, aberrant Wnt signaling transduction has a positive or negative influence on the cancer patient’s immune system cells. Tumor-intrinsic Wnt signaling has an impact on the immunogenicity of malignant cells. Some components of the WNT signaling pathway cascades that up-expression in malignancies cells could be recognized by immune cells as tumor-associated antigens (TAAs) such as the mutant b-catenin was recognized by autologous cytotoxic lymphocytes (CTLs) in melanoma (46). However, canonical or non-canonical Wnt signaling limits the functions of dendritic cells in achieving immunity tolerance in melanoma, and this process is associated with metabolic immunosuppression via vitamin A (47) and indoleamine 2,3-dioxygenase 1 (IDO1) (48). As the core members of the Wnt signal pathway, vaccination with DKK1 has been confirmed as the activator of CD4+ and CD8+ T lymphocytes in eliminating the myeloma cells in the murine model (49). However, the role of DKK1 in lung cancer has remained controversial. A number of studies have reported that the overexpression of DKK1 in NSCLC may lead to adverse outcomes. However, in the present study, the result of the meta-analysis did not show a significant difference in OS between aberrant DKK1 and normal DKK1 expression. This result is inconsistent with many studies. The most common aberrant type of DKK1 in lung cancer is missense not amplification. To our knowledge, missense cannot lead to gene overexpression, which suggests that the DKK1 expression levels might have been affected by other factors. Given this uncertainty, DKK1 function in lung cancer should be further investigated and discussed.
Taken together, the above results indicate that DKK1 has distinct functions and expression levels in different cancers, and further confirm that DKK1 can be a biomarker for prognosis, diagnosis, target therapy, and even perhaps, tumor vaccination. The tumorigenesis process cannot be clearly classified by a pathway or gene. Besides Wnt signaling, the PPI network analysis showed that DKK1 had many interactive genes including SMAD9, KREMN1-2, DKK2-DKK4, MDFI, LRP5-6, and others. The relation of DKK1 with these genes, especially the DKK family members, should be further explored to gain a better of understanding of DKK1’s functions in cancers. Our study used several online databases based on the most popular bioinformatics theories to perform a comprehensive analysis between the genes and tumors. The advantages of this method are its access to a massive sample population, lower cost, and large-scale genomic research and functional analysis capabilities. It provides a way to explore the target genes and the outcomes of cancer patients using large-scale online data. However, its online nature is also a limitation, and further verification through experiments is needed to confirm the findings of the present study. Further, we cannot construct a model to find the differential genes that have an impact on the prognosis of tumor.
Conclusions
DKK1, a member of the Dickkopf family, is as an inhibitor of Wnt signaling transduction. The differential expression of DKK1 is closely related to carcinogenesis, response to therapy, and metastasis. In this study, we performed a systemic analysis of the differential expression of DKK1 and explored the relation with the prognosis of defined cancer types. The results showed that DKK1 was significantly overexpressed in HNSC, LUSC, and PAAD, with the up-expression indicating poor outcome. Apart from the Wnt signaling pathway, DKK1 also interacts with the development and induction of other pathways.
This bioinformatics analysis revealed the prognostic and therapeutic value of DKK1 expression, contributed to clarifying its function in types of cancers. Based on this information, clinicians and researchers may develop potential target drugs for therapy in cancer patients and further explore the mechanisms underlying carcinogenesis of various types of cancers.
Acknowledgments
Funding: Innovation Project of Guangxi Graduate Education (No. YCSW2020118), Clinical Key Specialized Subject Construction Project of Guangxi Zhuang Autonomous Region (No. 2018-6).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3263). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDeri 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Diederich S, Wormanns D, Heindel W. Lung cancer screening with low-dose CT. Eur J Radiol 2003;45:2-7. [Crossref] [PubMed]
- Solanki S, Chakinala RC, Haq KF, et al. Inpatient burden of gastric cancer in the United States. Ann Transl Med 2019;7:772. [Crossref] [PubMed]
- White MC, Babcock F, Hayes NS, et al. The history and use of cancer registry data by public health cancer control programs in the United States. Cancer 2017;123:4969-76. [Crossref] [PubMed]
- Schootman M, Ratnapradipa K, Loux T, et al. Individual-and county-level determinants of high breast cancer incidence rates. Transl Cancer Res 2019;8:S323-33. [Crossref]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003;100:3983-8. [Crossref] [PubMed]
- Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res 2007;67:1030-7. [Crossref] [PubMed]
- Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994;367:645-8. [Crossref] [PubMed]
- Li L, Osdal T, Ho Y, et al. SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute myeloid leukemia stem cells. Cell Stem Cell 2014;15:431-46. [Crossref] [PubMed]
- Sadarangani A, Pineda G, Lennon KM, et al. GLI2 inhibition abrogates human leukemia stem cell dormancy. J Transl Med 2015;13:98. [Crossref] [PubMed]
- Miki T, Yasuda SY, Kahn M, et al. Wnt/β-catenin signaling in embryonic stem cell self-renewal and somatic cell reprogramming. Stem Cell Rev Rep 2011;7:836-46. [Crossref] [PubMed]
- Huang Y, Yang X, Zhao F, et al. Overexpression of Dickkopf-1 predicts poor prognosis for patients with hepatocellular carcinoma after orthotopic liver transplantation by promoting cancer metastasis and recurrence. Med oncol (Northwood, London, England) 2014;31:966.
- Yamabuki T, Takano A, Hayama S, et al. Dikkopf-1 as a novel serologic and prognostic biomarker for lung and esophageal carcinomas. Cancer Res 2007;67:2517-25. [Crossref] [PubMed]
- Takahashi N, Fukushima T, Yorita K, et al. Dickkopf-1 is overexpressed in human pancreatic ductal adenocarcinoma cells and is involved in invasive growth. Int J Cancer 2010;126:1611-20. [PubMed]
- Haaber J, Abildgaard N, Knudsen LM, et al. Myeloma cell expression of candidate genes for osteolytic bone disease - overexpression of DKK1 correlates with clinical bone involvement. Br J Haematol 2008;140:25-35. [PubMed]
- Seo HY. DKK1 overexpression might induce crizotinib resistance. AACR; Annual Meeting 2019; March 29-April 3, 2019; Atlanta, GA.
- Aufderklamm S, Hennenlotter J, Leidenberger P, et al. Systemic Alterations of Wnt Inhibitors in Patients with Prostate Cancer and Bone Metastases. Dis Markers 2018;2018:1874598.
- Gosepath EM, Eckstein N, Hamacher A, et al. Acquired cisplatin resistance in the head–neck cancer cell line Cal27 is associated with decreased DKK1 expression and can partially be reversed by overexpression of DKK1. Int J Cancer 2008;123:2013-9. [Crossref] [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucl Acids Res 2017;45:W98-102. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Liu SH, Shen PC, Chen CY, et al. DriverDBv3: a multi-omics database for cancer driver gene research. Nucl Acids Res 2020;48:D863-70. [PubMed]
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucl Acids Res 2010;38:W214-20. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cai L, Lin S, Girard L, et al. LCE: an open web portal to explore gene expression and clinical associations in lung cancer. Oncogene 2019;38:2551-64. [Crossref] [PubMed]
- Polakis P. Wnt signaling and cancer. Genes Develop 2000;14:1837-51. [PubMed]
- Mazieres J, He B, You L, et al. Wnt signaling in lung cancer. Cancer Lett 2005;222:1-10. [Crossref] [PubMed]
- Murillo-Garzón V, Kypta R. WNT signalling in prostate cancer. Nat Rev Urology 2017;14:683-96. [Crossref] [PubMed]
- Pohl S-G, Brook N, Agostino M, et al. Wnt signaling in triple-negative breast cancer. Oncogenesis 2017;6:e310. [Crossref] [PubMed]
- Teeuwssen M, Fodde R. Wnt Signaling in Ovarian Cancer Stemness, EMT, and Therapy Resistance. J Clin Med 2019;8:1658. [Crossref] [PubMed]
- Salim H, Zong D, Hååg P, et al. DKK1 is a potential novel mediator of cisplatin-refractoriness in non-small cell lung cancer cell lines. BMC Cancer 2015;15:628. [Crossref] [PubMed]
- Ogoshi K, Kasamatsu A, Iyoda M, et al. Dickkopf-1 in human oral cancer. Int J Oncol 2011;39:329-36. [PubMed]
- Takahashi N, Fukushima T, Yorita K, et al. Dickkopf‐1 is overexpressed in human pancreatic ductal adenocarcinoma cells and is involved in invasive growth. Int J Cancer 2010;126:1611-20. [PubMed]
- Han SX, Zhou X, Sui X, et al. Serum dickkopf-1 is a novel serological biomarker for the diagnosis and prognosis of pancreatic cancer. Oncotarget 2015;6:19907-17. [Crossref] [PubMed]
- Kimura H, Yamamoto H, Harada T, et al. CKAP4, a DKK1 receptor, is a biomarker in exosomes derived from pancreatic cancer and a molecular target for therapy. Clin Cancer Res 2019;25:1936-47. [Crossref] [PubMed]
- Gao H, Li L, Xiao M, et al. DKK1Elevated expression is an independent unfavorable prognostic indicator of survival in head and neck squamous cell carcinoma. Cancer Manag Res 2018;10:5083-9. [Crossref] [PubMed]
- Kuphal S, Lodermeyer S, Bataille F, et al. Expression of Dickkopf genes is strongly reduced in malignant melanoma. Oncogene 2006;25:5027-36. [Crossref] [PubMed]
- Wang S, Zhang S. Dickkopf-1 is frequently overexpressed in ovarian serous carcinoma and involved in tumor invasion. Clin Exp Metastasis 2011;28:581-91. [Crossref] [PubMed]
- Aguilera O, Fraga MF, Ballestar E, et al. Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene 2006;25:4116-21. [Crossref] [PubMed]
- Forget MA, Turcotte S, Beauseigle D, et al. The Wnt pathway regulator DKK1 is preferentially expressed in hormone-resistant breast tumours and in some common cancer types. Br J Cancer 2007;96:646-53. [Crossref] [PubMed]
- Fulciniti M, Tassone P, Hideshima T, et al. Anti-DKK1 mAb (BHQ880) as a potential therapeutic agent for multiple myeloma. Blood 2009;114:371-9. [Crossref] [PubMed]
- Linde N, Casanova-Acebes M, Sosa MS, et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat Commun 2018;9:21. [Crossref] [PubMed]
- Binda E, Visioli A, Giani F, et al. Wnt5a Drives an Invasive Phenotype in Human Glioblastoma Stem-like Cells. Cancer Res 2017;77:996-1007. [Crossref] [PubMed]
- Pukrop T, Klemm F, Hagemann T, et al. Wnt 5a signaling is critical for macrophage-induced invasion of breast cancer cell lines. Proc Natl Acad Sci U S A 2006;103:5454-9. [Crossref] [PubMed]
- Robbins PF, El-Gamil M, Li YF, et al. A mutated beta-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J Exp Med 1996;183:1185-92. [Crossref] [PubMed]
- Hong Y, Manoharan I, Suryawanshi A, et al. β-catenin promotes regulatory T-cell responses in tumors by inducing vitamin A metabolism in dendritic cells. Cancer Res 2015;75:656-65. [Crossref] [PubMed]
- Zhao F, Xiao C, Evans KS, et al. Paracrine Wnt5a-β-Catenin Signaling Triggers a Metabolic Program that Drives Dendritic Cell Tolerization. Immunity 2018;48:147-60.e7. [Crossref] [PubMed]
- Qian J, Zheng Y, Zheng C, et al. Active vaccination with Dickkopf-1 induces protective and therapeutic antitumor immunity in murine multiple myeloma. Blood 2012;119:161-9. [Crossref] [PubMed]