
Circulating tumor DNA correlates with microvascular invasion and predicts tumor recurrence of hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) remains a great threat to public health worldwide (1). Surgery is the mainstream modality that can offer a cure for HCC patients. However, postoperative tumor recurrence happens in more than 60% HCC patients within 5 years (2) and stands as the main obstacle for further prolonging the overall survival (OS) (3). Thus, it’s critical for clinical surgeons to predict patients with a high possibility of recurrence and apply suitable adjuvant therapy to improve the prognosis. Unfortunately, the current-existing clinical indices are hardly satisfactory in predicting tumor relapse. Being the most representative tumor marker, AFP has been serving to diagnose HCC and detect postoperative recurrence for decades, yet its sensitivity and specificity are still to be improved (4-6). Other markers such as PIVKA-II, AFP-L3 and Glypican-3 (GPC3) were also proved to be useful in HCC detection and surveillance (7,8), but they still served as complementary modalities to AFP (6). More importantly, the diagnostic value for recurrence of these markers was mostly effectuated by monitoring the fluctuating patterns (9) which called for continuous medical tests and was unlikely to be completed within perioperative periods. Therefore, novel method that can timely predict postoperative tumor recurrence is urgently needed.
In recent years, liquid biopsy has been greatly employed in the oncology research as a non-invasive and highly-sensitive technology (10). Specially, circulating free DNA or cell free DNA (cfDNA) has gained researchers’ interest and been used in the field of tumor prognosis, diagnosis and treatment choice (10-12). As an unneglected part of cfDNA and originated from tumor cells (13), circulating tumor DNA (ctDNA) has also been brought to clinical usage. It’s noteworthy that, ctDNA has been reported to predict tumor recurrence months ahead of image evidence (14), thus providing more chance for clinical intervention and prolonging OS. By far, the prognostic value of cfDNA or ctDNA was executed through concentration (15-19), which might be influenced by the extraction methods and patients’ physiological states (20-24). Previously, we have reported that ctDNA could be detected in HCC patients using hotspot mutants on the droplet digital PCR (ddPCR) platform. However, it’s still unknown whether ctDNA could predict tumor recurrence in HCC. Herein, for the first time, we systematically assayed the pre- and postoperative ctDNA in HCC patients using this ddPCR platform and found that positive ctDNA mutants correlated with microvascular invasion (MVI) and predicted tumor recurrence.
Methods
Patients’ enrollment
From March 2013 to July 2015, patients hospitalized in Zhongshan Hospital, Fudan University were enrolled based on the following criterion: (I) no previous history of malignancy; (II) no synchronous malignancies in other organs; (III) no anti-tumor treatments before hepatectomy; (IV) curative hepatectomy; (V) the pathological diagnosis of the tumor was HCC.
Sample collection and preparation
Ten milliliter EDTA tube (BD, Plymouth, UK) was used to collect blood drawn from ulnar vein. Blood samples were obtained in the morning following hospitalization and on the 7th postoperative day, respectively. A two-step centrifugation method was adopted to process the blood samples within 3 hours: (I) 10 minutes’ spin at 3,000 rpm to separate the blood cells; (II) another 10 minutes’ spin at 13,000 rpm for the removal of the cellular debris. The clarified plasma and PBMCs were stored at −80 °C for future ctDNA extraction. PBMCs were obtained from the blood cells centrifugated in the first step. Detailed methods of PBMCs and ctDNA acquiring were formulated in a previous research (25).
Sanger sequencing
DNA extracted from PBMCs was defined as germline DNA. Sequencing was performed after the germline DNA was amplified and purified. The processed DNA product was used in each reaction. The sequencing was carried out in parallel to ddPCR in order to avoid any cross-interference.
ddPCR
The ddPCR was carried out on the QX200 platform (Bio-Rad, Hercules, USA) according to the instructions. Firstly, 10 µL 2× ddPCR SuperMix (Bio-Rad), 1 µL forward and reverse primer (1:1 mixture), 1 µL probe for wild and mutant type (1:1 mixture), 3 µL deionized distilled water and 5 µL ctDNA template were blended into a 20 µL volume system. Secondly, the reaction system was jointly loaded to droplet generator together with 70 µL droplet generation oil to form 13,000 to 20,000 droplets. Then the droplets were transferred into a 96-well PCR plate in aliquots of 40 µL for amplification. The sequence information was presented in Table S1.
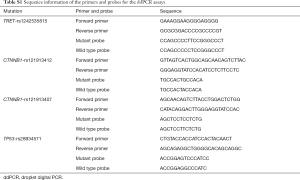
Full table
The PCR plate was placed in the Droplet Reader (Bio-Rad) after the amplification and was analyzed by QuantaSoft (Bio-Rad). Allele concentrations of mutant (CMUT, copies/µL) and wild type (CWT, copies/µL) were calculated and mutant allele frequency (MAF) was defined as CMUT/(CMUT + CWT), detailed PCR programmes and MAF calculation were described in a previous study (25).
Follow-ups
Follow-up evaluations were performed 1 month after surgery, then every 3 months within the first two years, every 6 months since the third postoperative year. Abdominal ultrasonography and levels of serum tumor markers (such as AFP) were regularly examined in every follow-up. Abdominal computed tomography (CT) or magnetic resonance imaging (MRI) was commenced if necessary. Any suspicious recurrence was confirmed or ruled out by an immediate abdominal enhanced MRI, chest CT or positron emission tomography (PET)/CT. Puncture biopsy was performed when the imaging diagnose was ambiguous. All patients were followed regularly till recurrence, death, or termination of the study. Disease free survival (DFS) and OS were used to represent the interval from the surgery to recurrence and from the surgery to death, respectively.
Statistical analysis
Statistical analysis was performed using SPSS 18.0 (IBM). Clinical parameters in different groups were compared using Chi-Square test. Cox regression model was established for univariate and multivariate analyses. The area under the curve (AUC) of receiver operating characteristic (ROC) curve was calculated to evaluate the diagnostic utility of ctDNA and MAF. The DFS and OS were compared by Kaplan-Meier curves. All P values were two-sided and P<0.05 was considered statistically significant.
Results
Patients’ demographics
Totally, 81 patients diagnosed with HCC were enrolled. The clinicopathological and epidemiological information were summarized in Table 1. The majority of the patients were male (69/81, 85.2%) with a median age of 54.7 (range, 28–78) years old. In all, 69 patients were HBsAg positive and 68 patients had liver cirrhosis. Before surgery, 46 patients had AFP level above 20 ng/mL, 61 patients’ Child-Pugh Scores were rated A and 43 patients were classified as BCLC (Barcelona Clinic Liver Cancer) stage (26) 0 or A while 38 patients were classified as B to C. The patients’ distributions according to the guidelines for primary liver cancer in China (27) were Ia [39], Ib [23], IIa [14] and IIb [5]. All patients successfully received hepatectomy with curative intent, and the Edmonson grades of their tumors were I–II [56] and III–IV [25].
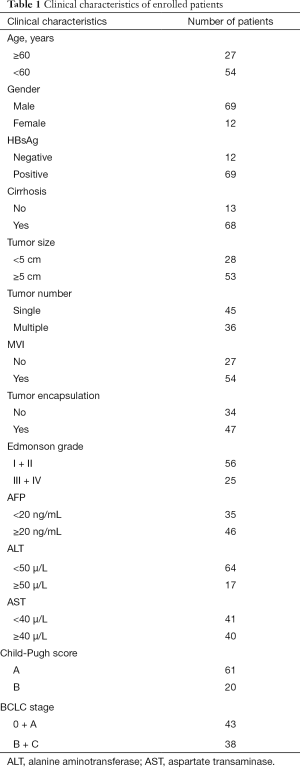
Full table
Determination of lower of detection (LOD) of ddPCR platform and baseline ctDNA status
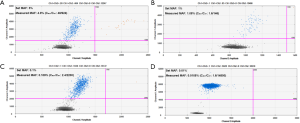
In our pilot research, we have established the LOD of QX200 platform by using serial dilution of KRAS G12D mutant (25). However, this mutant was not a frequent event in HCC. Herein, we further confirmed the platform’s working performance by additionally evaluating the LOD using the hotspot mutant TP53 rs28934571 of HCC. We compared the preset MAF with the measured MAF by ddPCR. As shown in Figure S1, the platform could stably detect the mutant allele at the MAF of 5%, 1%, 0.1% and 0.01% respectively, which was consistent with our previous result. Thus, the LOD in our study was 0.01%.
We then detected the four mutant alleles in the preoperative plasma DNA using this ddPCR platform. Totally, 57 patients (70.4%) were found to be ctDNA positive before surgery, of which 45 patients had 1 positive mutant allele, 11 patients had 2 positive alleles and 1 patient had 3 mutants. In general, the MAFs ranged from 0.02% to 43.28%, which were all above the LOD, thus ruling out the possibilities of false positivity. TP53 (c.747G>T) mutant was detected in 12 patients, TERT (c.1-124C>T) was found positive in 40 patients, and CTNNB1 (c.121A>G) and (c.133T>C) were presented in 12 and 8 patients, respectively. Also, we assayed the matched PBMCs for the corresponding hotspot mutations using sanger sequencing in order to exclude the possibility of germline mutation. In all the 81 patients, none of the mutants were detected in the germline DNA. Thus, the mutants detected in plasma DNA were genuinely ctDNA. Detailed mutation information of ddPCR was presented in Table S2.
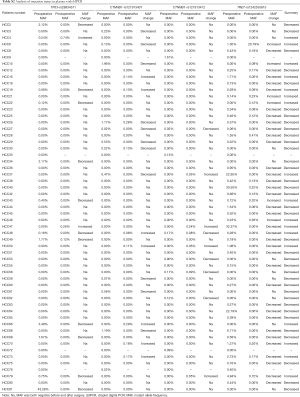
Full table
Preoperative ctDNA status correlates with prognosis
We then tried to analyze whether there was a correlation between preoperative ctDNA status and clinicopathological characteristics, and found that positive plasmatic ctDNA status before surgery significantly correlated with larger tumor size (P=0.001), more tumor lesions (P=0.001), more incidences of MVI (P<0.001), more advanced BCLC stages (P<0.001) and more events of tumor recurrence (P=0.018). Other parameters, such as cirrhosis, etiology, patient’s age did not significantly affect the detection rate of ctDNA (Table 2).
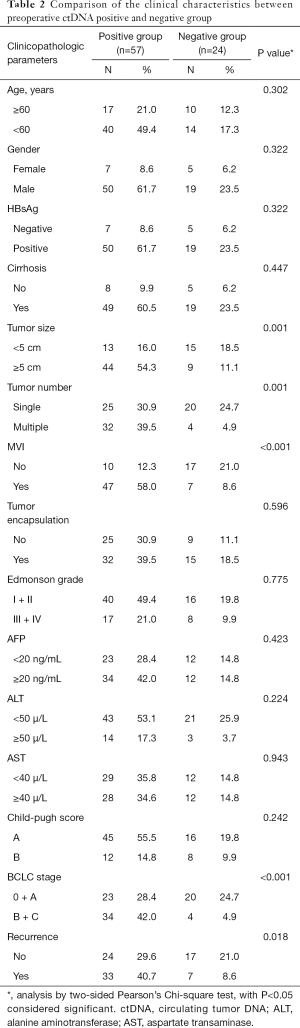
Full table
Then, we compared the DFS and OS between the ctDNA positive and negative groups using Kaplan-Meier curves. As shown in Figure 1, positive preoperative ctDNA status was associated with both shorter OS (mean OS 22.5 vs. 40.0 months, P<0.001, Figure 1A) and DFS (mean DFS 16.6 vs. 35.3 months, P<0.001, Figure 1B).
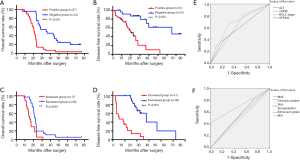
Next, cox regression models were established and the univariate and multivariate analyses were carried out to reveal the risk factors for tumor recurrence of the patients from the common clinical markers. We found that ctDNA status, high AFP level (AFP ≥400 ng/mL), ALT level and BCLC stage were the risk factors for postsurgical recurrence in univariate analysis. And ctDNA, ALT and BCLC stage were further proved to be the risk factors for recurrence by multivariate assessment (Table 3).
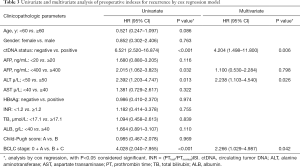
Full table
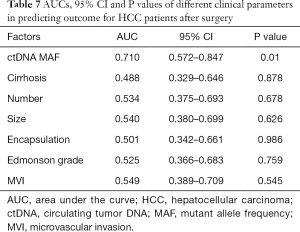
On the bias of the multivariate analysis, we generated ROC curves to evaluate the ability of each risk factor to predict postoperative tumor relapse in HCC patients (Figure 1E). The AUCs, 95% confident interval (CI) and P values were summarized in Table 4. Following BCLC stage, ctDNA ranked the second place in predicting postoperative recurrence among all the parameters assessed, with very close AUCs (0.625 vs. 0.675).
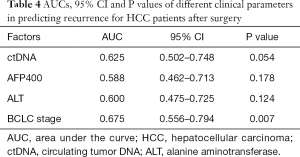
Full table
ctDNA dynamic changes predict tumor recurrence
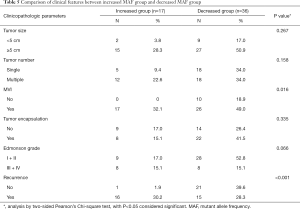
The postoperative plasma samples of 53 preoperative ctDNA positive patients were available and we then evaluated whether ctDNA still existed upon tumor resection. Considering intratumoral heterogeneity (ITH) may hinder comprehensive genomic profiling, we assayed all the four mutants rather than the positive allele before surgery. We found that ctDNA disappeared in 23 (23/53, 43.4%) patients and the MAF of ctDNA decreased in 13 (13/53, 24.5%) patients. In 17 (17/53, 32.1%) patients, the MAF of ctDNA increased or novel mutants that were not detectable before surgery were observed.
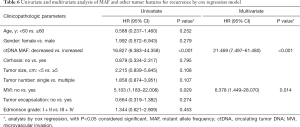
In order to evaluate the clinical significance of ctDNA dynamics, we classified the patients into two groups: the ones with negative mutant or decreased ctDNA MAFs were in decreased group while the others with novel mutant or increased ctDNA MAFs were in the increased group. The tumor features were compared between the two groups (Table 5). Patients with increased ctDNA MAF postoperatively had a markedly higher recurrence rate (P<0.001) and MVI positivity percentage (P=0.016) compared to those with decreased ctDNA MAF. Kaplan-Meier analysis showed that the mean OS was 16.8 months for the increased group and 25.3 months for the decreased group (P=0.0045, Figure 1C), while the DFS for the increased group was 7.0 nd 20.8 months for the decreased group (P<0.001, Figure 1D). We also performed cox regression analysis to reveal the correlated risk factors for postoperative recurrence in these patients. Both univariate and multivariate analyses showed that MVI and ctDNA MAF were the two independent risk factors for recurrence (Table 6) and ctDNA MAF possessed the greatest AUC for predicting postoperative recurrence in the ROC curves (Table 7 and Figure 1F).
Full table
Full table
Full table
Discussion
HCC ranks the sixth in morbidity and the fourth in mortality worldwide (28) while in China the fifth in both morbidity and cancer-related deaths (29). Due to the endemic hepatitis B virus (HBV) infection, Chinese population accounted for more than half the new cases of HCC (30). The features of multistep and multicentric carcinogenesis of HCC often result in cancer recurrence and poor prognosis (31). Therefore, early prediction and detection of HCC recurrence are critical in prolonging patients’ survival. However, even the most applied biomarker, AFP, with the cutoff value at 20 ng/mL, only had the sensitivity and specificity ranging from 41% to 65% and 80% to 94%, respectively (5). Therefore, novel biomarkers which could predict and detect HCC recurrence reliably and timely are urgently needed.
Recent years witnessed concentrated attentions on the studies of ctDNA. Harboring broad genetic information, ctDNA functions as a more comprehensive tool in analyzing tumoral genome compared to conventional sampling method (10). The relevant fields include unveiling drug resistance (11), monitoring treatment response (12), early diagnosis (32) and detection of incipient recurrence (14). Detailed research revealed close correlations between tumor recurrence and detectable postsurgical ctDNA as well as dynamic ctDNA changes (33,34).
Being one of the commonly used modalities that can overcome the low fraction ctDNA accounts for in cfDNA (35), ddPCR allows the detection of mutant down to 0.001%, which is superior to qPCR (36). In our study, ctDNA extracted from less than 5 mL peripheral blood plasma was sufficient for ddPCR reactions, this also highlighted the low demand of sample capacity (36). At the same time, ddPCR also provides a possibility of absolute quantification of nucleic acids since each reaction takes place in an individual droplet. However, considering the limited throughput expected from ddPCR and multi-genetic variations in HCC, a pre-time selection of targeted mutants was recommended. According to our previous studies, TP53, CTNNB1 and TERT were the most reported mutated genes in HCC (37-42). By furtherly testifying the relevance between mutant types and liver carcinogenesis, we narrowed the targeted mutants down to the up-mentioned four spots (25).
Our results showed that the preoperative mutant detection rate was 70.4%, while the positive rate of AFP was only 56.8%. By adding more relevant hotspots, the detection rate could be furtherly improved, amplifying the advantages of ctDNA. More importantly, detectable ctDNA before surgery was directly related to tumor size, tumor number, MVI and BCLC stage, which linked the mutants harbored in ctDNA with the tumor characteristics. This might be explained by the following reasons, since ctDNA is often considered fragments of tumor DNA in circulation, larger tumor size, more tumor lesions and vascular invasion by the tumor indicate higher possibilities of ctDNA release (43), which may result in more detections. The close connection between ctDNA and tumor features makes ctDNA an ideal potential tumor marker for recurrence. Both the ROC and OS/DFS assessments proved our speculation. Not only were the OS and DFS significantly shorter in the ctDNA positive group, the AUC of ctDNA status was also greater than AFP. Though the diagnostic value of ctDNA was topped by BCLC stage, considering the limited mutation spots, the performance of a more comprehensive and well-established mutant panel for ctDNA is worth waiting.
It is reported that the half-life time of ctDNA in circulation was no more than 2 hours (44); thus it’s reasonable to expect either a complete loss or a detectable decrease in MAF by the time we performed the second ddPCR and assume that the mutants detected in plasma 7 days after surgery were all from tumor cells still remaining in the liver, either as early intrahepatic metastasis or from the positive surgical margin. Apparently, disappearance of ctDNA or decrease of MAF was observed in most patients after resection, which should be the result of tumor removal. Still, we found increase or new-onset of mutant in some cases. We assume that this may be the result of the cancer biological behavior and ITH. Over half of the HCC patients still developed recurrence even with R0 resection and presence of minimally residual disease (MRD) was often considered to be responsible for that (45). These MRDs, being stimulated by surgical trauma, might function as the sources of the ctDNA after surgery, resulting in MAF increases. The close correlation between increased MAF and MVI furtherly proved our assumption. ITH played another key role in the MAF fluctuation. The vast amounts of mutants included within the HCCs laid the ground of great heterogeneity among the micro-lesions, causing possibilities of release of various kinds of hotspots from each focus, so that the ctDNA detected after surgery might harbor mutants different from the ones before surgery. Taken together, the detectable mutations in ctDNA after resection should be a solid evidence of MRD and indicate great chances of recurrence. This went along with our results. We found that patients with increased MAF manifested a significant shorter OS and DFS, moreover, the diagnostic value of MAF surpassed MVI, furtherly emphasizing the potential clinical application of ctDNA for recurrence.
Another interesting phenomenon was that we found that unlike the preoperative situation, the fluctuations of MAF and MVI were the only two factors remaining for recurrence, tumor size or tumor number failed to maintain the correlation. We speculated that this might be caused by the curative resections. Although larger tumor size and more tumor lesions might result in higher possibilities of recurrence (46), complete removal of the tumor could reduce the rate. By thorough radiographic examinations, both tumor size and tumor number can be acquainted with, therefore, specified resection strategies would be formulated to ensure a curative hepatectomy. On the other hand, MVI could only be diagnosed by postoperative pathology, there were no effective means to predict or handle MVI before hands. This reflected the superiority of MVI and ctDNA MAF in foretelling recurrence compared to the other tumor features.
According to our study, both the preoperative ctDNA status and fluctuations of ctDNA MAF performed well in predicting postoperative HCC relapse, still, there are some differences between the two indices. Taken more as a preoperative marker, ctDNA status could be used to predict tumor relapse after resection, its diagnostic ability was proved to be superior to that of AFP. The fluctuations of MAF, on the other hand, are more of a reflection of the surgery. Declined MAFs could be indicating a complete tumor resection while upgoing MAFs or novel mutant spots might be the sign of MRDs or micro-metastasis.
There are still some limitations in our study. First, this is a domestic research mainly concerning about Chinese population, the result should be furtherly validated in other group of people. Second, although we have observed associations between preoperative ctDNA positive status and several clinicopathological factors, it should be cautiously interpreted since we have only employed four mutants to detect ctDNA and this may result in false negativity, which would bias the association. To evaluate how ctDNA might be correlated with clinicopathological factors, deep sequencing of ctDNA with a panel of recurrently mutated genes in HCC would be more appropriate.
In conclusion, our study demonstrated a close correlation between plasma ctDNA and tumor features in HCC patients. Both preoperative ctDNA and dynamic fluctuations of MAF in ctDNA could be useful clinical markers for tumor relapse after surgery.
Acknowledgments
Funding: This study was jointly supported by the National Key R&D Program of China (2019YFC1315800, 2019YFC1315802), National Natural Science Foundation of China (No. 81830102, 81772578, 81802991, 81602581), STCSM (18YF1403600), and Shanghai Municipal Key Clinical Specialty.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for full data access, integrity of the data and the accuracy of the data analysis and ensure that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee (the Institutional Review Board of Zhongshan Hospital, Fudan University) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from every patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Bruix J, Sherman M. American Association for the Study of Liver D. Management of hepatocellular carcinoma: an update. Hepatology 2011;53:1020-2. [Crossref] [PubMed]
- Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378-90. [Crossref] [PubMed]
- Javadpour N. The role of biologic tumor markers in testicular cancer. Cancer 1980;45:1755-61. [Crossref]
- Lok AS, Sterling RK, Everhart JE, et al. Des-gamma-carboxy prothrombin and alpha-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology 2010;138:493-502. [Crossref] [PubMed]
- Cui R, He J, Zhang F, et al. Diagnostic value of protein induced by vitamin K absence (PIVKAII) and hepatoma-specific band of serum gamma-glutamyl transferase (GGTII) as hepatocellular carcinoma markers complementary to alpha-fetoprotein. Br J Cancer 2003;88:1878-82. [Crossref] [PubMed]
- Volk ML, Hernandez JC, Su GL, et al. Risk factors for hepatocellular carcinoma may impair the performance of biomarkers: a comparison of AFP, DCP, and AFP-L3. Cancer Biomark 2007;3:79-87. [Crossref] [PubMed]
- Gao F, Zhu HK, Zhu YB, et al. Predictive value of tumor markers in patients with recurrent hepatocellular carcinoma in different vascular invasion pattern. Hepatobiliary Pancreat Dis Int 2016;15:371-7. [Crossref] [PubMed]
- Kim DY, Paik YH, Ahn SH, et al. PIVKA-II is a useful tumor marker for recurrent hepatocellular carcinoma after surgical resection. Oncology 2007;72 Suppl 1:52-7. [Crossref] [PubMed]
- Luke JJ, Oxnard GR, Paweletz CP, et al. Realizing the potential of plasma genotyping in an age of genotype-directed therapies. J Natl Cancer Inst 2014.106. [PubMed]
- Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [Crossref] [PubMed]
- Montagut C, Siravegna G, Bardelli A. Liquid biopsies to evaluate early therapeutic response in colorectal cancer. Ann Oncol 2015;26:1525-7. [Crossref] [PubMed]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [Crossref] [PubMed]
- Reinert T, Scholer LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016;65:625-34. [Crossref] [PubMed]
- Agassi R, Czeiger D, Shaked G, et al. Measurement of circulating cell-free DNA levels by a simple fluorescent test in patients with breast cancer. Am J Clin Pathol 2015;143:18-24. [Crossref] [PubMed]
- da Silva Filho BF, Gurgel AP, Neto MA, et al. Circulating cell-free DNA in serum as a biomarker of colorectal cancer. J Clin Pathol 2013;66:775-8. [Crossref] [PubMed]
- Nygaard AD, Holdgaard PC, Spindler KL, et al. The correlation between cell-free DNA and tumour burden was estimated by PET/CT in patients with advanced NSCLC. Br J Cancer 2014;110:363-8. [Crossref] [PubMed]
- Singh N, Gupta S, Pandey RM, et al. High levels of cell-free circulating nucleic acids in pancreatic cancer are associated with vascular encasement, metastasis and poor survival. Cancer Invest 2015;33:78-85. [Crossref] [PubMed]
- Chen K, Zhang H, Zhang LN, et al. Value of circulating cell-free DNA in diagnosis of hepatocelluar carcinoma. World J Gastroenterol 2013;19:3143-9. [Crossref] [PubMed]
- Page K, Guttery DS, Zahra N, et al. Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS One 2013;8:e77963. [Crossref] [PubMed]
- Mazurek AM, Fiszer-Kierzkowska A, Rutkowski T, et al. Optimization of circulating cell-free DNA recovery for KRAS mutation and HPV detection in plasma. Cancer Biomark 2013;13:385-94. [Crossref] [PubMed]
- Page K, Powles T, Slade MJ, et al. The importance of careful blood processing in isolation of cell-free DNA. Ann N Y Acad Sci 2006;1075:313-7. [Crossref] [PubMed]
- Sozzi G, Roz L, Conte D, et al. Effects of prolonged storage of whole plasma or isolated plasma DNA on the results of circulating DNA quantification assays. J Natl Cancer Inst 2005;97:1848-50. [Crossref] [PubMed]
- Ramachandran K, Speer CG, Fiddy S, et al. Free circulating DNA as a biomarker of prostate cancer: comparison of quantitation methods. Anticancer Res 2013;33:4521-9. [PubMed]
- Huang A, Zhang X, Zhou SL, et al. Detecting Circulating Tumor DNA in Hepatocellular Carcinoma Patients Using Droplet Digital PCR Is Feasible and Reflects Intratumoral Heterogeneity. J Cancer 2016;7:1907-14. [Crossref] [PubMed]
- Bruix J, Sherman M. Practice Guidelines Committee AAftSoLD. Management of hepatocellular carcinoma. Hepatology 2005;42:1208-36. [Crossref] [PubMed]
- Zhou J, Sun HC, Wang Z, et al. Guidelines for Diagnosis and Treatment of Primary Liver Cancer in China (2017 Edition). Liver Cancer 2018;7:235-60. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Tang ZY. Hepatocellular carcinoma surgery--review of the past and prospects for the 21st century. J Surg Oncol 2005;91:95-6. [Crossref] [PubMed]
- Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem 2015;61:112-23. [Crossref] [PubMed]
- Sausen M, Phallen J, Adleff V, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun 2015;6:7686. [Crossref] [PubMed]
- Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133. [Crossref] [PubMed]
- Mouliere F, Robert B, Arnau Peyrotte E, et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS One 2011;6:e23418. [Crossref] [PubMed]
- Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604-10. [Crossref] [PubMed]
- Fujimoto A, Totoki Y, Abe T, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet 2012;44:760-4. [Crossref] [PubMed]
- Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012;44:694-8. [Crossref] [PubMed]
- Huang J, Deng Q, Wang Q, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet 2012;44:1117-21. [Crossref] [PubMed]
- Cleary SP, Jeck WR, Zhao X, et al. Identification of driver genes in hepatocellular carcinoma by exome sequencing. Hepatology 2013;58:1693-702. [Crossref] [PubMed]
- Kan Z, Zheng H, Liu X, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res 2013;23:1422-33. [Crossref] [PubMed]
- Schulze K, Imbeaud S, Letouze E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505-11. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Kustanovich A, Schwartz R, Peretz T, et al. Life and death of circulating cell-free DNA. Cancer Biol Ther 2019;20:1057-67. [Crossref] [PubMed]
- Erstad DJ, Tanabe KK. Prognostic and Therapeutic Implications of Microvascular Invasion in Hepatocellular Carcinoma. Ann Surg Oncol 2019;26:1474-93. [Crossref] [PubMed]
- Tokumitsu Y, Sakamoto K, Tokuhisa Y, et al. A new prognostic model for hepatocellular carcinoma recurrence after curative hepatectomy. Oncol Lett 2018;15:4411-22. [PubMed]


