
Screening TCGA database for prognostic genes in lower grade glioma microenvironment
Introduction
Glioma is neoplasm of the central nervous system (CNS), derived from transformed neural stem cells or progenitor cells (1). According to histopathological characteristics, the World Health Organization (WHO) classifies glioma into two categories: low-grade glioma (LGG, grade I and II), which is well differentiated and slow-growing; and high-grade glioma (HGG, grade III and grade IV), which is poorly differentiated or anaplastic, and most of them seriously infiltrate the brain parenchyma. LGG is a fatal disease that predisposes to young people (mean age 41 years), with an average survival time of about 7 years, and all LGGs will eventually develop into HGG (2). Interestingly, in addition to conventional surgical treatment, LGG has limited sensitivity to radiation and chemotherapy (3-6). Since immune checkpoint therapies such as nivolumab have developed rapidly in LGG in recent years, tumor microenvironment (TME) has attracted increasing attention as a crucial cellular milieu for immune cells, stromal cells and extracellular matrix molecules (7,8).
TME is a cellular environment in which neoplastic foci are located. It consists of endothelial cells, inflammatory mediators, mesenchymal cells, along with immune cells and stromal cells (8). Among them, immune cells and stromal cells are two major non-tumor components, which are of great significance in the diagnosis and prognosis of cancers. LGG tumor cells form a complex milieu of the tumor microenvironment, which ultimately promotes the adaptability of the transcriptome of tumor cells and disease progression (8). On the other hand, studies have shown that TME can have an important impact on tumor malignancy and clinical prognosis by regulating gene expression (9-14).
To further investigate the molecular biological properties of TME, algorithms that use gene expression data of The Cancer Genome Atlas (TCGA) database, which is a comprehensive genome-wide gene expression profile established to classify and detect genomic abnormalities in a large number of populations worldwide, have been developed (12,15-17). For example, an algorithm called ESTIMATE that uses expression data to estimate stromal cells and immune cells in malignant tumors has been designed by Yoshihara et al. (12). In this algorithm, the expression characteristics of specific genes in immune cells and stromal cells are analyzed to calculate immune and stromal scores in order to predict the invasion of non-tumor cells. Recent reports showed that ESTIMATE was applied in researches of prostate cancer (18), breast cancer (19) and colon cancer (20). However, characteristics of TME evaluated by ESTIMATE were not observed in LGG.
To get more insights, we extracted a list of microenvironment-related genes that predicted poor prognosis in patients of LGG by using the TCGA database of LGG cohorts and the immune scores derived from ESTIMATE algorithm (12), what is more important, we developed a risk scoring system to assess the prognostic value of central genes. In addition, the correlation between central genes and immune infiltration has also been explored.
Methods
TCGA and ESTIMATE data collection
We downloaded gene transcriptome profiles of patients with LGG of the central nervous system in The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). RNA expression for LGG Multiforme was obtained by using IlluminaHiSeq (version: 2017-10-13). After that, we download the survival data through the Genomic Data Commons (GDC) tool from TCGA. Sex, histological type and tumor grade were extracted. Limma package was used for normalization processing (21). Scores of immune, stromal and ESTIMATE were calculated using ESTIMATE algorithm (https://sourceforge.net/projects/estimateproject/).
Correlation analysis and survival analysis
We performed unpaired t test and ordinary one-way analysis of variance to explore the association between immune scores, stromal scores, ESTIMATE scores and histological type, tumor grade. Kaplan-Meier (K-M) analysis with log-rank test was based on survival package (22,23). P value <0.05 was considered as statistically significant in mentioned analyses.
Heatmaps, clustering analysis and DEGs
Immune scores and stromal scores were divided into high-level groups and low-level groups according to the median scores, respectively. Limma package with the standard of |log(FC)| >1 and False Discovery Rate (FDR) <0.05 were used for standardization of transcriptome data (21). Heatmaps (|log(FC)| >1 and FDR <0.05) and volcano plots (cut |log2FC|=1 and cut P=0.05) based on pheatmap package, ggplot2 package and clustering analysis were applied for visualizing expression of differential expressed-gene screening and cluster analysis. We obtained the intersected differentially expressed genes (DEGs) among immune scores and stromal scores which exhibited by VennDiagram package (24).
DEGs enrichment analysis and GSEA
Online tool DAVID (The Database for Annotation, Visualization and Integrated Discovery, https://david.ncifcrf.gov/) was exploited to construct the gene ontology (GO) categories by biological processes (BP), cellular components (CC) and molecular functions (MF) (25). Besides, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was performed based on org.Hs.eg.db package, clusterProfiler, org.Hs.eg.db, enrichplot, and ggplot2 packages. The q-value <0.05 was considered as significance. In addition, ‘c2.cp.kegg.v6.2.symbols.gmt gene sets’ was set as gene set database, ‘Illumina_Human.chip’ was set as chip platform, FDR <0.25, |enriched score| >0.35, and gene size ≥35 during Gene Set Enrichment Analysis (GESA) process (26).
PPI network construction and hub genes selection
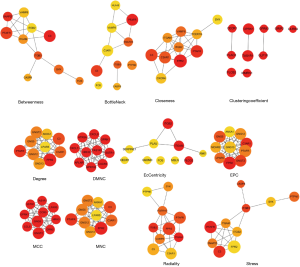
We constructed protein-protein interaction (PPI) network from STRING database (version 11.0, minimum required interaction score: 0.9) and Cytoscape software (version 3.7.1) (27,28). We used the inside-software plugin cytoHubba for hub genes identification and top 10 nodes ranked by every algorithm were enrolled in gene selection (29). Genes with network nodes less than 10 were excluded. Circle size represented node degree.
Overall survival curve and risk score
K-M analysis was used to evaluate the prognostic value of hub genes. Log-rank test was used and P value <0.05 was considered as statistically significant. Furthermore, we established a risk score (RS)-based prognostic evaluation model, which formula is RS=Ʃ (βi*Expi). In this formula, ‘i’ represented the number of prognostic hub genes, ‘Expi’ represented the selected gene, and ‘βi’ represented the weight of gene, which calculated based on multivariate cox regression. Then we calculated RS of LGG patients and divided them into high- and low-risk groups according to the median RS. Survival receiver operating characteristic curve (ROC) package was used (30). In addition, K-M plots were used to evaluate the prognostic value of RS.
TIMER database analysis
In tumor immune estimation resource (TIMER) database (https://cistrome.shinyapps.io/timer/), RNA-Seq expression profiling data were utilized to detect the infiltration of immune cells including B cells, CD4+ T cells, CD8+ T cells, Neutrphils, Macrophages and Dendritic cells in tumor tissues. Besides, the association between tumor purity and hub genes was also analyzed.
Statistical analysis
IBM SPSS Statistics 20.0 was applied in multivariate Cox regression analysis and K-M analysis. Statistical analysis was implemented on R software (version 3.5.2). The results with P<0.05 represented as statistically significant.
Results
Immune scores and stromal scores are associated with LGG sub-types, tumor grade and survival outcome
Totally 530 LGG tumor data were collected from TCGA. Then, we screened a total of 516 samples ended with ‘-01’, which represented ‘primary solid tumor’. Among them, we deleted samples TCGA-CS-5390-01 and TCGA-R8-A6YH-01, because we were unable to obtain survival information for these samples. Finally, 514 LGG samples including 285 males (55.45%) and 229 females (44.55%) were extracted from TCGA database. Clinical characters of LGG patients were exhibited in Table 1. Immune scores, stromal scores and ESTIMATE scores were listed in online: http://cdn.amegroups.cn/static/application/954ad2d4809e5d6e033eaf346ed86465/atm.2020.01.73-t1.pdf. The distribution of immune scores, stromal scores and ESTIMATE scores in different histological types and grades was shown in Figure 1A,B,C,D,E,F. The results revealed that higher scores were significantly related with histological types and higher tumor grade (P<0.0001, respectively). Besides, K-M curves revealed that higher scores were associated with worse survival outcomes in immune scores (P=0.0167), stromal scores (P=0.0035) and ESTIMATE scores (P=0.0190) (Figure 1G,H,I).
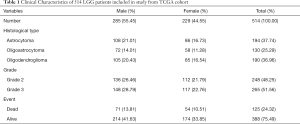
Full table
Comparison of gene expression profiles with immune scores and stromal scores in LGG
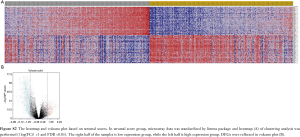
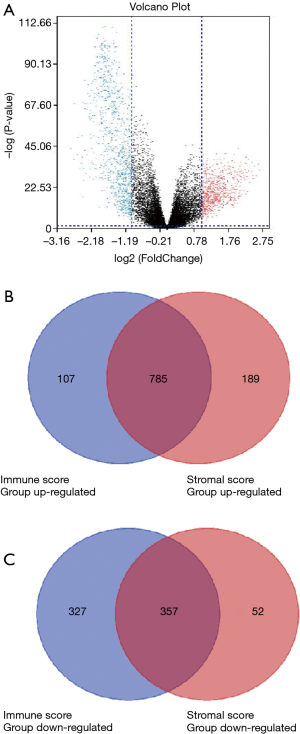
Microarray data was standardized by limma package. In immune score group, heatmap of clustering analysis was shown in Figure S1. DEGs were reflected in volcano plot (Figure 2A). The heatmap and volcano plot based on stromal scores were shown in Figure S2A,B. According to statistics, 684 up-regulated DEGs and 892 down-regulated DEGs were selected in immune score group (online: http://cdn.amegroups.cn/static/application/09167c99881221da5e479a7b81a141af/atm.2020.01.73-t2.docx), while 409 up-regulated DEGs and 974 down-regulated DEGs were selected in stromal score group (online: http://cdn.amegroups.cn/static/application/4847a27dac216bb9faa00192b15e5428/atm.2020.01.73-t3.docx). Then, 785 up-regulated intersected genes and 357 down-regulated intersected genes were revealed in venn plots (Figure 2B,C).

Functional enrichment analysis
The results of GO analysis revealed that 1142 intersected genes were associated with immune response, inflammatory response, plasma membrane and receptor activity from the categories of BP (Figure 3A), CC (Figure 3B) and MF (Figure 3C), respectively. KEGG pathway annotation was analyzed and shown in Figure 3D. We separately calculated the enrichment of intersected genes in the aspects of organismal systems, human diseases, environmental information processing, cellular process metabolism and genetic information processing. From the results, we found that intersected genes were most abundant in pathways of the immune system, infectious diseases, signal transduction, signaling molecules and interaction, cell transport and catabolism, gloable and overview maps and folding sorting and degradation. Besides, KEGG enrichment barplot was shown in Figure 3E. Top 20 pathway enrichment analysis was visually presented in Figure 3F, which bubble size represented gene number and colour represented Q value.
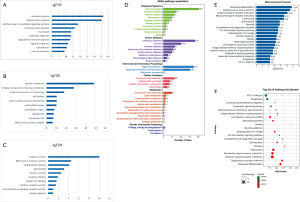
GSEA
To explore the difference in the pathway between high and low immune score groups, we performed GSEA analysis and the results were shown in Figure S3A, FcγR mediated phagocytosis, leukocyte transendothelial migration, natural killer cell mediated cytotoxicity, cell adhesion molecules, B cell receptor signaling pathway, Toll like receptor signaling pathway, JAK-STAT signaling pathway, cytokine-cytokine receptor interaction, T cell receptor signaling pathway, FcεRI signaling pathway, NOD like receptor signaling pathway, intestinal immune network for IgA production, antigen processing and presentation were intersected genes enriched pathways. In addition, Figure S3B shown the results of GSEA analysis conducted on stromal scores groups. Allograft rejection, antigen processing and presentation, viral myocarditis, autoimmune thyroid disease, cell adhesion molecules, graft versus host disease, intestinal immune network for IgA production, JAK-STAT signaling pathway, leishmania infection, NOD like receptor signaling pathway, systemic lupus erythematosus, Toll like receptor signaling pathway were intersected genes enriched pathways.
PPI
PPI network was performed via STRING tool for exploring the interplay among the identified DEGs. The network contained 1,103 nodes and 4,070 edges. Then, data from STRING were further analyzed by Cytoscape and hub genes identification was performed by cytoHubba, which results of algorithms were shown in Figure S4. The 25 tumor immune-related hub genes were identified as follow: HRH3, APLNR, FCER1G, SYK, GNG12, GNG13, GNG5, PTPN6, GPR183, CXCL11, CXCL9, HLA-B, VAMP8, SSTR1, CCR5, PTAFR, C3, TAS1R1, ANXA1, OPRK1, ITGB2, CCL5, CXCR3, CXCR4 and GNGT2.
RS and survival analysis
Based on the selected hub genes, RS=HRH3*(−0.045)+APLNR*0.063+FCER1G*0.03+SYK*(−0.535)+GNG12*0.269+GNG13*0.024+GNG5*0.344+PTPN6*(−0.523)+GPR183*0.346+CXCL11*0.181+CXCL9*(−0.13)+HLA-B*(−0.026)+VAMP8*(−0.302)+SSTR1*(−0.174)+CCR5*0.093+PTAFR*0.23+C3*(−0.37)+TAS1R1*0.171+ANXA1*0.158+OPRK1*0.147+ITGB2*0.381+CCL5*−0.371+CXCR3*0.245+CXCR4*(−0.119)+GNGT2*0.597.
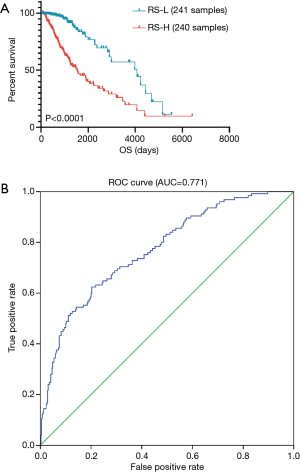
We selected 481 patients with OS >30 days from 514 samples. Then RS data of 481 patients were calculated and shown in online: http://cdn.amegroups.cn/static/application/e64541dfb3b1cd2b8e9900fe0c982188/atm.2020.01.73-t4.pdf. The range of RS was −3.99 to 3.03 and the median was −0.5. Totally 481 eligible LGG patients were divided into low-RS group and high-RS group according to the median RS. Survival analysis was constructed and the result demonstrated that patients with higher RS were associated with worse overall survival than who with lower RS (P<0.0001, Figure 4A). Furthermore, ROC curve was used to evaluate the prognostic value of RS. AUC was calculated as 0.771, which revealed superior predictive accuracy in overall survival (Figure 4B). In addition, survival curves of 25 hub genes were shown in Figure S5. From the K-M curves, lower expression levels of GNG13, HRH3, OPRK1 and SSTR1 were related with poor prognosis. In contrast, we found that high expression of other hub genes is associated with poor prognosis in LGG patients.

Hub genes and immune cells infiltration
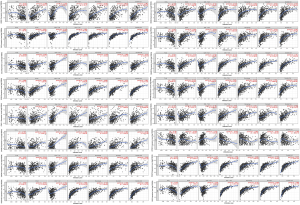
TIMER database was applied for further exploring the relationship between 25 hub genes and immune infiltration in LGG TME. According to the results, 16 hub genes (ANAX1, C3, CCL5, CCR5, CXCL11, CXCR3, CXCR4, FCER1G, GNG5, GNG12, GNGT2, GPR183, HLA-B, OPRK1, PTAFR, SYK) were related with B cell, CD4+T cell, CD8+T cell, macrophage, neutrophil and dendritic cell infiltration (P<0.05, respectively, Figure S6).
Discussion
In recent years, due to the rapid development of immunological checkpoint therapies such as nivolumab in LGG, TME has attracted a growing number of attentions as a key cellular milieu of immune cells, extracellular matrix molecules and stromal cells (7,8). Immunotherapy for cancers destroys cancer cells by enhancing the function of immune system. Growing evidence indicates that the key mechanism of interaction between the immune system and LGG is the immune checkpoints in the dynamic effect of immunity. Defined as a cohort of co-stimulatory and co-inhibitory molecules modulating T-cell activity, immune checkpoints orchestrate as regulatory circuits to make the immune system self-tolerable under normal physiological circumstances (31,32).
In our current research, we calculated the immune scores, stromal scores and ESTIMATE scores for each LGG sample extracted from the TCGA database by applying ESTIMATE algorithm. The results revealed that immune scores were statistically higher in malignant tumor cases and associated with worse survival outcomes, advanced tumor grades and higher pathological stages. For the first time that ESTIMATE algorithm-derived immune scores were calculated in LGG to evaluate the prognostic value and provide extra evidence for the biological basis of immunotherapy. In our study, PPI network was constructed using SRING tool and Cytoscape software. Finally, 25 TME-related hub genes were selected and the potential pathways such as immune response, inflammatory response, plasma membrane and receptor activity were identified. We explored the associations between hub genes with immune infiltration in LGG TME by using the deconvolution algorithm based on the TIMER database. We found 16 hub genes including CCL5, CCR5, CXCR3, CXCR4 and CXCL11 were related to B cell, CD4+T cell, CD8+T cell, macrophage, neutrophil and dendritic cell infiltration.
Chemokine (C-C motif) ligand 5 (CCL5) is secreted by various cell types including platelets, immune cells, fibroblasts, and endothelial and epithelial cells (33), and has been originally identified as an inducer that can recruit leukocytes to sites of inflammation (34). After binding to its receptors, namely CCR1, CCR3, and CCR5, CCL5 induces phosphorylation of mitogen-activated protein kinase and other signaling pathways involved in the regulation of various cellular functions, such as the proliferation, migration, and differentiation (35,36). Interestingly, it has been reported that the CCL5/CCR1 axis is critical for maintaining mesenchymal stem cells (MSC) identity and multipotency (37). Non-neoplastic cell-derived signals (chemokines and cytokines) in the TME may also represent viable treatment targets. It was reported that CCL5 was responsible for maintaining neurofibromatosis type 1 (NF1) mouse optic glioma growth in vivo (38). Since malignant gliomas may achieve partial independence from growth regulatory factors produced by non-neoplastic cells in the TME by producing the same cytokines secreted by the stromal cells in their low-grade counterparts, the NF1 protein, neurofibromin, negatively regulates CCL5 expression through suppression of AKT/mTOR signaling (39). Besides, CCL5 operates through an unconventional CCL5 receptor, CD44, to inhibit mouse mesenchymal glioblastoma (M-GBM) cell apoptosis. Collectively, these findings reveal that paracrine factors important for LGG growth can be usurped by high-grade tumors to create autocrine regulatory circuits that maintain malignant glioma survival (40). The opinions provided possible ideas for further research on our results.
The C-X-C motif chemokine receptor 3 (CXCR3), which belongs to the CXC chemokine receptor family, is a G protein-coupled receptor that plays a critical role in mediating chemotactic migration, cell proliferation, and survival (41-43). CXCR3 is mainly expressed by various effector T lymphocytes including CD4+ Th1 cells, CD8+ cytotoxic T cells, and natural killer (NK) cells (44,45). The principle chemokine ligands of CXCR3 are CXCL4, CXCL9, CXCL10 and CXCL11 (46-49). Maru et al. (50) first reported that CXCR3 and CXCL10 were increased in glioma cells compared with adult human astrocytes. Moreover, they found that the expression level of CXCR3 correlated with malignancy grade of glioma. Liu et al. (51) examined the role of CXCR3 in glioblastoma (GBM) progression using the GL261 murine model of malignant glioma. They found that Murine glioma GL261 cells express CXCL10 in vitro and GL261 tumors express CXCL9 and CXCL10 in vivo. CXCR3−/− mice with glioma had significantly shorter median survival time and reduced numbers of tumor-infiltrated natural killer (NK) and natural killer T (NKT) cells in contrast with WT glioma mice. Moreover, both CXCR3 and its ligands are expressed by murine and human glioma cell lines (A172, T98G, U87, U118 and U138). These results suggested that CXCR3 system may be a unique target for human glioma therapy. Recently, Pu et al. (52) investigated the potential prognostic value of CXCR3 in primary GBM and its relationship with clinicopathological features. Using K-M survival curve analysis with a log-rank comparison of the 65 primary glioma patients, they found that the patients with higher expression levels of CXCR3 had shorter progression free survival and overall survival compared with those with lower expression levels of CXCR3. Using univariate Cox regression analysis, they found that high CXCR3 expression was a risk factor for primary GBM [P<0.01, hazard ratio (HR) 2.336; 95% confidence interval (CI), 1.341–4.071]. Furthermore, their multivariate Cox regression analysis showed that CXCR3 expression level was an independent prognostic factor for overall survival of primary glioma patients. However, the role of CXCR3 in LGG has not yet been elucidated. The results mentioned above are consistent with our analysis, suggesting that CXCR3 may be a useful independent prognostic biomarker for patients with primary glioma.
Remarkably, the risk model was calculated based on 25 hub prognostic genes associated with LGG TME. The AUC of the ROC curve revealed the satisfactory predictive efficiency of the risk signature. This novel TME hub genes-related risk score model provides a new theoretical basis for the prognosis assessment of LGG patients, which is expected to be further applied in the future clinical management. The interplay of LGG and its TME critically affects tumor evolution, which subsequently impacts subtype classification, recurrence, drug resistance, and the overall prognosis of patients. Previous reports have provided elegant analysis on how the activation of tumor-intrinsic genes shapes TME (53). In the current work, we focused on genes characteristic of microenvironment, which in turn affect the development of LGG and hence contribute to patients’ overall survival. Our results may provide additional data in decoding the complex interaction of tumor and tumor environment in LGG.
It is important to note that limitations existed in our current study. Firstly, we only selected target data from the TCGA public database through biological algorithm approaches. We should validate the results of this article in clinical patients in further study. Secondly, 16 hub genes related to immune cells infiltration should be further studied to clarify the regulatory mechanism in immune infiltrates of LGG. Finally, considering the choice of analytical approaches, we included a limited database for the screening of hub genes in the immune ecosystem, which may lead to biased results due to the neglect of other databases.
Conclusions
In our research, we selected the transcriptional profiles from public databases based on bioinformatics algorithm and identified specific signatures that related to the infiltration of stromal and immune cells in LGG TME.
Acknowledgements
Funding: This work was supported by grants from the National Natural Science Foundation of China (Nos. 81871873 and 81570186), Jiangsu Province Key Research Development Program (BE2016794 to J Feng), Research Project of Jiangsu Cancer Hospital (ZM201801, ZN201602, ZM201908).
Footnote
Conflicts of Interest: XL serves as an unpaid editorial board member of Annals of Translational Medicine from Oct 2019 to Sep 2020. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ilkhanizadeh S, Lau J, Huang M, et al. Glial progenitors as targets for transformation in glioma. Adv Cancer Res 2014;121:1-65. [Crossref] [PubMed]
- Claus EB, Walsh KM, Wiencke JK, et al. Survival and low-grade glioma: the emergence of genetic information. Neurosurg Focus 2015;38:E6. [Crossref] [PubMed]
- Yan Y, Xu Z, Dai S, et al. Targeting autophagy to sensitive glioma to temozolomide treatment. J Exp Clin Cancer Res 2016;35:23. [Crossref] [PubMed]
- Dai S, Yan Y, Xu Z, et al. SCD1 Confers Temozolomide Resistance to Human Glioma Cells via the Akt/GSK3beta/beta-Catenin Signaling Axis. Front Pharmacol 2018;8:960. [Crossref] [PubMed]
- Yan Y, Li Z, Zeng S, et al. FGFR2-mediated phosphorylation of PTEN at tyrosine 240 contributes to the radioresistance of glioma. J Cell Commun Signal 2019;13:279-80. [Crossref] [PubMed]
- Forst DA, Nahed BV, Loeffler JS, et al. Low-grade gliomas. Oncologist 2014;19:403-13. [Crossref] [PubMed]
- Escudier B, Sharma P, McDermott DF, et al. CheckMate 025 Randomized Phase 3 Study: Outcomes by Key Baseline Factors and Prior Therapy for Nivolumab Versus Everolimus in Advanced Renal Cell Carcinoma. Eur Urol 2017;72:962-71. [Crossref] [PubMed]
- Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett 2017;387:61-8. [Crossref] [PubMed]
- Curry JM, Sprandio J, Cognetti D, et al. Tumor microenvironment in head and neck squamous cell carcinoma. Semin Oncol 2014;41:217-34. [Crossref] [PubMed]
- Cooper LA, Gutman DA, Chisolm C, et al. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am J Pathol 2012;180:2108-19. [Crossref] [PubMed]
- Galon J, Pages F, Marincola FM, et al. The immune score as a new possible approach for the classification of cancer. J Transl Med 2012;10:1. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Şenbabaoğlu Y, Gejman RS, Winer AG, et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol 2016;17:231. [Crossref] [PubMed]
- Winslow S, Lindquist KE, Edsjo A, et al. The expression pattern of matrix-producing tumor stroma is of prognostic importance in breast cancer. BMC Cancer 2016;16:841. [Crossref] [PubMed]
- Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30:413-21. [Crossref] [PubMed]
- Aldape K, Zadeh G, Mansouri S, et al. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol 2015;129:829-48. [Crossref] [PubMed]
- Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061-8. [Crossref] [PubMed]
- Shah N, Wang P, Wongvipat J, et al. Regulation of the glucocorticoid receptor via a BET-dependent enhancer drives antiandrogen resistance in prostate cancer. Elife 2017.6. [PubMed]
- Priedigkeit N, Watters RJ, Lucas PC, et al. Exome-capture RNA sequencing of decade-old breast cancers and matched decalcified bone metastases. JCI Insight 2017.2. [PubMed]
- Alonso MH, Ausso S, Lopez-Doriga A, et al. Comprehensive analysis of copy number aberrations in microsatellite stable colon cancer in view of stromal component. Br J Cancer 2017;117:421-31. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Aalen OO. A linear regression model for the analysis of life times. Stat Med 1989;8:907-25. [Crossref] [PubMed]
- Aalen OO. Further results on the non-parametric linear regression model in survival analysis. Stat Med 1993;12:1569-88. [Crossref] [PubMed]
- Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 2011;12:35. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Tilford CA, Siemers NO. Gene set enrichment analysis. Methods Mol Biol 2009;563:99-121. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8 Suppl 4:S11. [Crossref] [PubMed]
- Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 2000;56:337-44. [Crossref] [PubMed]
- Driessens G, Kline J, Gajewski TF. Costimulatory and coinhibitory receptors in anti-tumor immunity. Immunol Rev 2009;229:126-44. [Crossref] [PubMed]
- Syn NL, Teng MWL, Mok TSK, et al. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol 2017;18:e731-41. [Crossref] [PubMed]
- Lin SJ, Chang KP, Hsu CW, et al. Low-molecular-mass secretome profiling identifies C-C motif chemokine 5 as a potential plasma biomarker and therapeutic target for nasopharyngeal carcinoma. J Proteomics 2013;94:186-201. [Crossref] [PubMed]
- Donlon TA, Krensky AM, Wallace MR, et al. Localization of a human T-cell-specific gene, RANTES (D17S136E), to chromosome 17q11.2-q12. Genomics 1990;6:548-53. [Crossref] [PubMed]
- Aldinucci D, Colombatti A. The inflammatory chemokine CCL5 and cancer progression. Mediators Inflamm 2014;2014:292376. [Crossref] [PubMed]
- Marques RE, Guabiraba R, Russo RC, et al. Targeting CCL5 in inflammation. Expert Opin Ther Targets 2013;17:1439-60. [Crossref] [PubMed]
- Mohr A, Zwacka R. The future of mesenchymal stem cell-based therapeutic approaches for cancer - From cells to ghosts. Cancer Lett 2018;414:239-49. [Crossref] [PubMed]
- Solga AC, Pong WW, Kim KY, et al. RNA Sequencing of Tumor-Associated Microglia Reveals Ccl5 as a Stromal Chemokine Critical for Neurofibromatosis-1 Glioma Growth. Neoplasia 2015;17:776-88. [Crossref] [PubMed]
- Johannessen CM, Reczek EE, James MF, et al. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci U S A 2005;102:8573-8. [Crossref] [PubMed]
- Pan Y, Smithson LJ, Ma Y, et al. Ccl5 establishes an autocrine high-grade glioma growth regulatory circuit critical for mesenchymal glioblastoma survival. Oncotarget 2017;8:32977-89. [PubMed]
- Ślusarczyk J, Trojan E, Chwastek J, et al. A Potential Contribution of Chemokine Network Dysfunction to the Depressive Disorders. Curr Neuropharmacol 2016;14:705-20. [Crossref] [PubMed]
- Van Raemdonck K, Van den Steen PE, Liekens S, et al. CXCR3 ligands in disease and therapy. Cytokine Growth Factor Rev 2015;26:311-27. [Crossref] [PubMed]
- Loetscher M, Gerber B, Loetscher P, et al. Chemokine receptor specific for IP10 and mig: structure, function, and expression in activated T-lymphocytes. J Exp Med 1996;184:963-9. [Crossref] [PubMed]
- Altara R, Manca M, Brandao RD, et al. Emerging importance of chemokine receptor CXCR3 and its ligands in cardiovascular diseases. Clin Sci (Lond) 2016;130:463-78. [Crossref] [PubMed]
- Cao F, Chen SS, Yan XF, et al. Evaluation of side effects through selective ablation of the mu opioid receptor expressing descending nociceptive facilitatory neurons in the rostral ventromedial medulla with dermorphin-saporin. Neurotoxicology 2009;30:1096-106. [Crossref] [PubMed]
- Cole KE, Strick CA, Paradis TJ, et al. Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J Exp Med 1998;187:2009-21. [Crossref] [PubMed]
- Lasagni L, Francalanci M, Annunziato F, et al. An alternatively spliced variant of CXCR3 mediates the inhibition of endothelial cell growth induced by IP-10, Mig, and I-TAC, and acts as functional receptor for platelet factor 4. J Exp Med 2003;197:1537-49. [Crossref] [PubMed]
- Farber JM. A macrophage mRNA selectively induced by gamma-interferon encodes a member of the platelet factor 4 family of cytokines. Proc Natl Acad Sci U S A 1990;87:5238-42. [Crossref] [PubMed]
- Luster AD, Unkeless JC, Ravetch JV. Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature 1985;315:672-6. [Crossref] [PubMed]
- Maru SV, Holloway KA, Flynn G, et al. Chemokine production and chemokine receptor expression by human glioma cells: role of CXCL10 in tumour cell proliferation. J Neuroimmunol 2008;199:35-45. [Crossref] [PubMed]
- Liu C, Luo D, Reynolds BA, et al. Chemokine receptor CXCR3 promotes growth of glioma. Carcinogenesis 2011;32:129-37. [Crossref] [PubMed]
- Pu Y, Li S, Zhang C, et al. High expression of CXCR3 is an independent prognostic factor in glioblastoma patients that promotes an invasive phenotype. J Neurooncol 2015;122:43-51. [Crossref] [PubMed]
- Wang Q, Hu B, Hu X, et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017;32:42-56.e6. [Crossref] [PubMed]


