A review of the application of deep learning in medical image classification and segmentation
Introduction
Since 2006, deep learning has emerged as a branch of the machine learning field in people’s field of vision. It is a method of data processing using multiple layers of complex structures or multiple processing layers composed of multiple nonlinear transformations (1). In recent years, deep learning has made breakthroughs in the fields of computer vision, speech recognition, natural language processing, audio recognition and bioinformatics (2). Deep learning has been praised as one of the top ten technological breakthroughs since 2013 due to its considerable application prospects in data analysis. The deep learning method simulates the human neural network. By combining multiple nonlinear processing layers, the original data is abstracted layer by layer, and different levels of abstract features are obtained from the data and used for target detection, classification or segmentation. The advantage of deep learning is to replace the manual acquisition feature with unsupervised or semi-supervised feature learning and hierarchical feature extraction efficient algorithms (3).
Medical care is about the health of people. At present, the amount of medical data is huge, but it is crucial to make good use of this huge medical data to contribute to the medical industry. Although the amount of medical data is huge, there are still many problems: medical data is diverse, including maps, texts, videos, magnets, etc.; due to different equipment used, the quality of data varies greatly; data presents fluctuating characteristics, over time and specific events change; due to differences in individuals, the law of the disease has no universal applicability (4). There are many factors that cannot be dealt with in the existence of these problems. Medical imaging is a very important part of medical data.
This paper first introduces the application of deep learning algorithms in medical image analysis, expounds the techniques of deep learning classification and segmentation, and introduces the more classic and current mainstream network models. Then we detailed the application of deep learning in the classification and segmentation of medical images, including fundus, CT/MRI tomography, ultrasound and digital pathology based on different imaging techniques. Finally, it discusses the possible problems and predicts the development prospects of deep learning medical imaging analysis.
Deep learning architectures
Deep learning algorithms
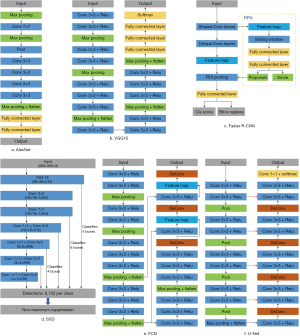
Deep learning has developed into a hot research field, and there are dozens of algorithms, each with its own advantages and disadvantages. These algorithms cover almost all aspects of our image processing, which mainly focus on classification, segmentation. Figure 1 is an overview of some typical network structures in these areas.
Classification
Using deep learning for image classification is earliest rise and it also a subject of prosperity. Among them, convolutional neural network (CNN) is the most widely structure. Since Krizhevsky et al. proposed AlexNet based on deep learning model CNN in 2012 (5), which won the championship in the ImageNet image classification of that year, deep learning began to explode. In 2013, Lin et al. proposed the network in network (NIN) structure, which uses global average pooling to reduce the risk of overfitting (6). In 2014, GoogLeNet and VGGNet both improved the accuracy on the ImageNet dataset (7,8). GoogLeNet has further developed the v2, v3 and v4 versions to improve performance (9-11). For the shortcomings of CNN on the input size fixed requirements, He et al. proposed spatial pyramid pooling (SPP) model to enhance the robustness of the input data (12). With the deepening of the deep learning model, He et al. proposed the residual network ResNet for the problem of model degradation that may occur, and continue to advance the deep learning technology (13).
Take AlexNet as an example. In 2012, the AlexNet adopted an 8-layer network structure consisting of five convolutional layers and three fully connected layers. After each convolution in five convolutional layers, a maximum pooling performed to reduce the amount of data. AlexNet accepts 227×227 pixels’ input data. After five rounds of convolution and pooling operations, the 6×6×256 feature matrix finally sent to the fully connected layer. The sixth layer of the fully connected layer sets up 4,096 convolution kernels, and the linear feature value of 4,096 size obtained by the dropout operation. After the last two layers, we get 1,000 float types output data, which is the final prediction result. AlexNet’s error rate in ImageNet was 15.3%, which was much higher than the 26.2% in second place. At the same time, its activation function is not sigmoid but adopted ReLU, and proved that the ReLU function is more effective.
VGG16 first proposed by VGG Group of Oxford University. Compared with AlexNet, it uses several consecutive 3×3 kernels instead of the larger convolution kernel in AlexNet like 11×11 and 5×5. For a given receptive field range, the effect of using several small convolution kernels is better than using a larger convolution kernel, because the multi-layer nonlinear layer can increase the network depth to ensure more complex patterns are learned, and the computational cost is also more small.
GoogLeNet, which launched in the same year as VGGNet, also achieved good results. Compared to VGGNet, GoogLeNet designed a module called inception. It’s a dense structure with a small number of convolution kernels of each size, and use 1×1 convolutional layer to reduce the amount of computation.
Segmentation
Semantic segmentation is an important research field of deep learning. With the rapid development of deep learning technology, excellent semantic segmentation neural networks emerge in large numbers and continuously become state-of-the-art in various segmentation competitions. Since CNN’s success in the classification field, people started to try CNN for image segmentation. Although CNN can accept images of any size as input, CNN will lose some details while pooling for extracting features, and it will lose the space information of input image due to the fully connected layers at the end of the network. So it’s difficult for CNN to pinpoint which category certain pixels belong to. With the development of deep learning technology, some segmentation networks based on convolution structure are derived.
The fully convolutional network (FCN) (14) proposed by Long et al. is the originator of the semantic segmentation networks. It replaces the fully connected layers of the classification network VGG16 with convolutional layers and retains the spatial information of the feature map and achieves pixel-level classification. Finally, FCN uses the deconvolution and fusing feature maps to restore the image, and provides the segmentation result of each pixel by softmax. Since the fully connected layer with dense connections is replaced by the convolutional layer which is locally connecting and weights sharing, the FCN greatly reduces the parameters that need to be trained. The performance of the FCN on the Pascal VOC 2012 datasets (15) has increased by 20% compared to the previous method, reaching 62.2% of the mIOU.
U-Net (16) was proposed by Olaf based on FCN, and has been widely used in medical imaging. Based on the idea of FCN deconvolution to restore image size and feature, U-Net constructs the encoder-decoder structure in the field of semantic segmentation. The encoder gradually reduces the spatial dimension by continuously merging the layers to extract feature information, and the decoder portion gradually restores the target detail and the spatial dimension according to the feature information. Among them, the step of the encoder gradually reducing the image size is called downsampling, and the step of the decoder gradually reducing the image details and size is called upsampling. Different from the fusion operation of the direct addition feature when the FCN is upsampled, the U-Net upsampling process first uses the concatenate operation to splicing the feature maps before the up-sampling of the encoder and the downsampling of the decoder. After concatenation the feature map is deconvolved. Different from the conventional convolution, pooling, and other operations, this strategy of directly utilizing shallow features is called skip connection. U-Net adopts the skip connection strategy of splicing to make full use of the features of the downsampling part of the encoder to be used for upsampling. To achieve a more refined reduction, this strategy is applied to shallow feature information of all scales to achieve a better reduction effect.
SegNet (17) is a depth semantic segmentation network designed by Cambridge to solve autonomous driving or intelligent robots, which is also based on the encoder-decoder structure. SegNet’s encoder and decoder each have 13 convolution layers. The convolutional layer of the encoder corresponds to the first 13 convolutional layers of VGG16. The upsampling part of the decoder uses UnPooling. SegNet records the element position information of the maximum pooling operation when the encoder is downsampled, and restores the image according to the position information when sampling on the decoder. SegNet with this strategy does not require learning when upsampling, and SegNet training is more accurate and faster than FCN.
In order to fuse the context information under multi-scale at the same level, PSPNet (18) proposes a pooled pyramid structure, which realizes image segmentation in which the target environment can be understood, and solves the problem that FCN cannot effectively deal with, the relationship problem between global information and scenes. Its pooled pyramid structure can aggregate context information of different regions, thereby improving the ability to obtain global information.
Deep learning development framework
While the deep learning technology is developing in theory, the software development framework based on deep learning theory is also booming.
Convolutional architecture for fast feature embedding (Caffe)
Caffe was born in Berkeley, California and now hosted by BVLC. Caffe features high-performance, seamless switching between CPU and GPU modes, and cross-platform support for Windows, Linux and Mac. Caffe has three basic atomic structures of Blobs, Layers and Nets, and its programming framework is implemented under these three atoms. It highly abstracts the structure of the deep neural network in terms of the “Layer”, and significantly optimizes the execution efficiency through some elaborate design, and it has flexibility based on maintaining efficient implementation.
Tensorflow
TensorFlow is an open source software library that uses data flow diagrams for numerical calculations. Google officially opened the computing framework TensorFlow on November 9, 2015, and officially released Google TensorFlow version 1.0 in 2017, marking its official use in the production environment. The TensorFlow calculation framework can well support various algorithms for deep learning such as CNN, RNN and LSTM, but its application is not limited to deep learning, but also supports the construction of general machine learning. TensorFlow’s components are excellent, and it provides powerful visualization capabilities through TensorBoard, which can generate very powerful visual representations of real-world network topologies and performance. At the same time, it supports heterogeneous distributed computing, which can run on multiple GPUs at the same time, and can automatically run the model on different platforms. Because TensorFlow developed in C++, it has high-performance.
PyTorch
Pytorch is the python version of torch, a neural network framework that is open sourced by Facebook and specifically targeted at GPU-accelerated deep neural network programming. Unlike Tensorflow’s static calculation graph, Pytorch’s calculation graph is dynamic, and the calculation graph can be changed in real-time according to the calculation needs. In January 2017, the Facebook Artificial Intelligence Institute (FAIR) team opened up PyTorch on GitHub and quickly occupied the top of the GitHub hotlist. PyTorch immediately attracted widespread attention as soon as it was launched, and quickly became popular in research.
High-performance computing based on GPU
The key factors of image processing in medical imaging field are imaging speed, image size and resolution. Due to the limitation of the hardware, the processing of medical images calculated according to sequence. It is also due to the lack of computing resources that the processing of these images wastes a lot of valuable time of doctors and patients. In recent years, GPU has made great progress and moved towards the direction of general computing. Its data processing capacity far exceeds that of CPU, which makes it possible to realize high-performance computing on ordinary computers.
The full name of the GPU is the Graphics Processing Unit, a microprocessor that performs image computing on PCs, workstations, game consoles and some mobile devices. In August 1999, NVIDIA released a GeForce 256 graphics chip codenamed NV10. Its architecture is very different from that of the CPU. At the beginning of its birth, it was mainly oriented to the rendering of graphic images. Like the CPU, the GPU is a processor in the graphics card that designed to perform complex mathematical and geometric calculations that are required for graphics rendering. With the GPU, CPU does not need to perform graphics processing work, and can perform other system tasks, which can greatly improve the overall performance of the computer.
Deep learning for medical imaging analysis
With the development of deep learning, computer vision uses a lot of deep learning to deal with various image problems. Medical image as a special visual image has attracted the attention of many researchers. In recent years, various types of medical image processing and recognition have adopted deep learning methods, including fundus images, endoscopic images, CT/MRI images, ultrasound images, pathological images, etc. At present, deep learning technology is mainly used in classification and segmentation in medical images. Figure 2 shows the main medical application scenarios of deep learning.
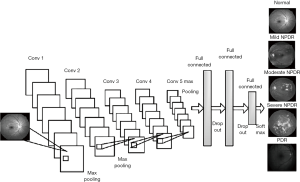
The classification of medical image
Diabetic retinopathy detection
In the field of deep learning, image classification and its application have made great progress this year. On the one hand, the academic circles have made great efforts to design a variety of efficient CNN models, which have achieved high accuracy and even exceeded the human recognition ability. On the other hand, the application of CNN model in medical image analysis has become one of the most attractive directions of deep learning. In particular, the retinal fundus image obtained from fundus camera has become one of the key research objects of deep learning in the field of image classification.
The main method for studying related fundus diseases using deep learning techniques is to classify and detect fundus images, such as diabetic retinopathy detection and glaucoma detection. The following Table 1 lists the deep learning methods applied and fundus image analysis in the past 3 years. These methods mainly use the large scale dataset to train deep CNN model and perform disease classification detection on fundus images. The deep CNN used to update iterations with the development of deep learning techniques, from the earliest shallow CNN model to the deep CNN model or some combination models, and the use of migration learning, data augmentation and other new methods and techniques.

Full table
We mainly work of detecting fundus diseases in transfer learning. It’s very difficult to obtain large-scale medical annotation set, and transfer learning is an effective method to solve the problem of small data. In order to find the potential factors between the accuracy and the type of primary models, we process transfer learning using the pre-trained model, for example CaffeNet, GoogleNet, VGG19. The experimental results show that the transfer learning based on the pre-trained CNN model is introduced to solve the problems in medical image analysis, and some effective results are produced. The Figure 3 shows our classification model of fundus.
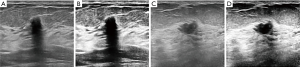
Ultrasound detection of breast nodules
The current deep learning technology has achieved research results in the field of ultrasound imaging such as breast cancer, cardiovascular and carotid arteries. Compared with traditional machine learning, deep learning can automatically filter features to improve recognition performance based on multi-layer models. Deep learning has become an important tool for ultrasonic image recognition with its high efficiency and accuracy, which can effectively improve the diagnostic accuracy. In the field of ultrasound imaging of breast nodules, Chen et al. combined the detection methods of texture features and morphology (25,26), using AlexNet model to find the nodules in the image and predicting the benign and malignant, the AUC value reached 0.9325 (27). Shalev-Shwartz et al. applied deep learning to the segmentation of the left ventricular ultrasound image of the heart, and achieved a better segmentation effect. In the study of automatic classification of fetal facial ultrasound images, Yu integrated CNN and random two-coordinate descent optimization algorithm (28), achieving 96.98% accuracy. At the same time, Yu uses the ResNet model in the automatic recognition of melanoma in dermoscopic images based on deep aggregation features (29,30). The AUC has reached more than 80%.
In the classification of breast nodules, we mainly propose an enhancement method of data preprocessing, called adaptive contrast enhancement (ACE) method. Directly sending the original data to the neural network training often has a poor effect. On this basis, we combine the ACE algorithm to enhance the ultrasound image. The ACE algorithm can calculate the relative pixel value of the target point and surrounding pixels by difference to correct the final pixel value, which has a good enhancement effect on the image. Figure 4 shows the enhanced image of breast.
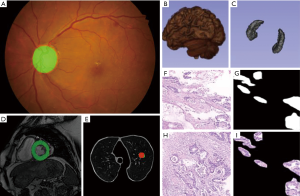
Pulmonary nodule screening
Pulmonary nodule disease is a common lung disease. Figure 5 shows the whole picture of pulmonary nodule in CT image. The accuracy rate of common X-ray chest film in the diagnosis of pulmonary nodules is less than 50%, and even people with normal chest film can be detected to infer sarcoidosis. With CT becoming the main detection method of pulmonary nodule diagnosis, more and more physical examinations begin to include lung cancer screening detection. According to relevant data statistics, the detection rate of pulmonary nodule has increased 5 times in recent years. With the development of deep learning technology, a series of deep learning methods are emerging to detect pulmonary nodules.
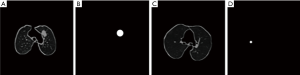
We compared a variety of existing lung nodule detection methods and found that the deep learning algorithm greatly improved the detection rate of nodules compared with the traditional machine learning algorithm. On the other hand, the deep learning algorithm also has its bottleneck in the detection of pulmonary nodules. First, the high detection rate of lung nodules depends on a large number of data sets, and at the same time depends on the accuracy of the annotation data. Secondly, as the number of network layers increases, the accuracy rate is not very large, which indicates that the deep learning algorithm itself has its limitations.
The segmentation of medical image analysis
Early detection of Alzheimer’s disease (AD)
Brain MRI analysis is mainly for the segmentation of different brain regions and the diagnosis of brain diseases, such as brain tumor segmentation (31), schizophrenia diagnosis, early diagnosis of Parkinson’s syndrome (32) and early diagnosis of AD. Among them, the broadest field of deep learning applications is the early diagnosis of AD. AD diagnosis based on deep learning is mainly based on segmentation of hippocampus, cortical thickness and brain volume in brain MRI images. Sarraf et al. (33) trained AD samples for sMRI and fMRI using the well-known LeNet-5 framework in CNNs, respectively, yielding 98.84% and 96.85% accuracy, respectively. This is the first time based on deep learning to analyzing fMRI data. Payan et al. (34) proposed a 3D CNN for AD diagnosis based on SAE pre-training. The 3D MRI scan is randomly selected to pre-train the SAE, and the trained SAE weights are used for 3D-CNN convolution filtering pre-training. Finally, the 3D-CNN fully connected layer is fine-tuned, but fine-tuning needs to be at the expense of a large amount of computational complexity during the training phase. Hosseini-Asl et al. (35) and other analysis based on sMRI feature extraction technology limits the accuracy of AD classification, they proposed a new depth-supervised adaptive 3D-CNN network, in which 3D-CAE learning and automatic extraction Identify AD features and capture changes caused by AD. The 3D-CAE pre-trained convolution filter is further applied to another set of data fields, such as the CAD Dementia pre-trained AD neuroimaging (ADNI) data set. In the early diagnosis of AD, our research group also carried out related experiments and proposed a deep learning method based on enhanced AlexNet. According to the characteristics of AD, we focus on the how to precise the hippocampus. We compared the segmentation of hippocampus with mainstream deep learning networks, including FCN, Unet, SegNet, Unet-3Dand Mask-RCNN. Figure 6 shows the segmented result of these networks.
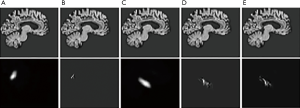
Furthermore, we use the dice coefficient as a measure. From the Figure 7, we can see that Mask-RCNN achieves high precision in hippocampus segmentation. At the same time, we noticed that the effect of Unet is far less than the effect of Unet-3D. The reason may be that MRI imaging itself is a three-dimensional form, so the use of three-dimensional convolution can better interpret the segmented object.
Left ventricular segmentation
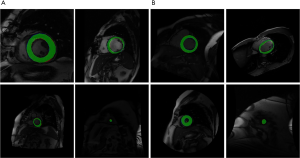
Cardiac MRI analysis diagnoses heart disease by dividing the left ventricle to measure left ventricular volume, ejection fraction, and wall thickness. Among them, deep learning is widely used in left ventricular segmentation. In recent years, deep learning algorithms for left ventricular segmentation on MRI image have emerged in an endless stream. Poudel et al. (36) proposed a recurrent full convolutional network (RFCN) that learns image representation from the entire 2D slice stack and utilizes inter-slice spatial dependence through internal memory cells, which can detect and segment anatomical Combined into a single architecture for end-to-end training, it significantly reduces computation time, simplifies the segmentation process, and enables real-time applications. Isensee et al. (37) proposed a method for classifying into a fully automated processing pipeline by integrating segmentation and disease, which uses a set of U-Net structures to segment the cardiac structure at each point in the cardiac cycle. Liao et al. (38) designed a detector combined with a neural network classifier to detect the ROI containing LV. The LV in the ROI is then segmented using a deep full CNN called hypercolumns. The 2D segmentation results are integrated into different images to estimate the volume. This model uses end-to-end training and uses the real volume directly as a label. Lieman-Sifry et al. (39) developed the FastVentricle architecture based on the ENet architecture. The FastVentricle architecture is a FCN architecture for ventricular partitioning that runs four times faster than the best ventricular partitioning structure and six times less memory. Maintain good clinical accuracy.
In this field, our group proposed a precise left cardiac contour segmentation model based on group normalization and nearest neighbor interpolation, which is called GNNI U-Net. We constructed a convolution module based on group normalization method for fast and accurately feature extraction, and an up-sampling module for feature restoring based on nearest neighbor interpolation method. Our model got Dice coefficient of 0.937 on the Sunnybrook dataset and 0.957 and on the LVSC dataset, which achieves high precision on left cardiac contour segmentation. Figure 8 shows our left ventricular segmentation effect.
Gastric cancer pathological
Pathological diagnosis is the “gold standard” of various cancer diagnosis methods, which plays an important role in the medical field (40). The contradiction between a large number of market demands and a shortage of talents in the field of pathology gives a great opportunity for deep learning in this field, and digital pathology technology also makes computer-assisted diagnosis possible (41).
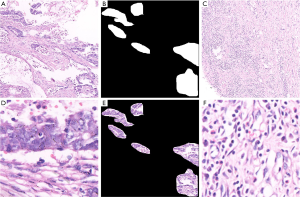
Nowadays, deep learning technology has been applied to the pathological diagnosis of lung cancer, breast cancer and gastric cancer. Its application mainly includes early tumor screening and benign and malignant diagnosis of tumor. In the field of lung cancer pathology, Zhang et al. established the “early computer diagnosis system for lung cancer” to detect lung cancer pathological sections, so that several major types of lung cancer can be detected (42). In the field of breast cancer pathology, Qaiser et al. established a CNN-based intelligent image diagnosis system with a judgment result of pathological sections (cancer and non-cancer tissue) as high as 0.833, four cancer classifications (normal tissue, benign tumor, original The accuracy of the results of both cancer and invasive cancer is as high as 0.778, which can reach the level of pathologists (43). Hanna and others based on the deep learning artificial intelligence automatic human epidermal growth factor receptor scoring system to score human epidermal growth factor receptor, the results are better than pathologists (44). An intelligent image diagnosis system constructed by Ehteshami Bejnordi et al. The diagnosis of gastric cancer pathological images can reach 69.9% accuracy compared with pathologists (45), Yoshida et al. reached 0.556 in three classifications of gastric cancer pathology (normal tissues, adenomas, cancer cells) (46). In the field of gastric cancer pathology our research team has established a benign and malignant diagnostic system based on gastric cancer pathology based on deep learning, with a sensitivity of over 97% (47). However, the above methods include that the initial work of our group rarely involves the field of image segmentation. In fact, the segmentation of pathological images is indeed a problem. We further designed a multi-input model called MIFNet to segment the lesions in the pathological image, and increase the dice coefficient to 81.87% in the segmentation of gastric cancer case images, much higher than some existing segmentation models. Such as U-Net (67.73%), SegNet (63.89%) and PSPNet (60.51%). Figure 9 shows some sample experimental data. What we need to do is get the label of the style shown in Figure 9B, which marks the tissue area of the lesion on the image, as shown in Figure 9E.
Conclusions and future research
Deep learning is one of the powerful tools for medical image analysis. It has been successfully applied in target detection, segmentation, classification and registration. The development of deep learning in the medical field depends on the accumulation of medical big data, while the medical data itself has multi-modal characteristics, which provides a large amount of rich data for deep learning. In terms of disease treatment, deep learning can not only find the lesion area, but also discriminate and classify specific lesions. When the lesion is positive, many detection networks can also segment the lesion area. While deep learning reflects its own advantages, it still has certain shortcomings. The deep learning model relies heavily on data sets. Each deep learning network requires massive data for training, which makes the data set acquisition more demanding. The root cause is that the pixel features from the original input image are too complex, so it is a future development trend to focus on designing a network with a smaller data size.
Deep learning widely applies all aspects of medical image analysis, including ophthalmology, neuroimaging, ultrasound, etc. With the development of deep learning, more and more medical fields will apply deep learning technology, and future deep learning will not only focus on the single aspect of neuroimaging but also other aspects of genomics and bioinformatics.
The reason why deep learning can develop so rapidly in the medical field is inseparable from a large number of clinical practices. How to better apply deep learning to all stages of medical treatment becomes a more challenging task. It depends on two aspects: one is the constantly updated iteration of technology, and the other is the continuous accumulation of medical experience.
At present, a number of excellent algorithms have emerged in the fields of driverlessness, natural language processing, computer vision, etc. These algorithms have attracted great attention in their respective fields, and how to use these advanced deep learning algorithms is an aspect worthy of our researchers’ constant thinking and innovation.
Deep learning on the medical imaging applications is not limited to the detection of big data routine diseases, but also effective solutions for rare diseases. At present, we mainly use the method of migration learning for small sample data to converge and achieve prediction results. However, there are certain limitations in migration learning. Not all rare diseases can be predicted in this way, which brings new challenges and opportunities for the diagnosis of intractable diseases.
With the advent of the 5G era, it provides a new and broad space for medical deep learning. Our traditional machine learning algorithms are concentrated at the software level, and there is very little technology involved in deep learning at the hardware level. The combination of 5G technology and deep learning technology enables the machine to achieve true intelligence. At the same time, the continuous development of intelligent medical devices and medical robots promotes the realization of deep learning at the hardware level, and greatly facilitates patient treatment.
Acknowledgments
Funding: None.
Footnote
Contributions: (I) Conception and design: J Gao, D Zhao; (II) Administrative support: J Gao; (III) Provision of study materials or patients: None; (IV) Collection and assembly of data: All authors; (V) Data analysis and interpretation: All authors; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors.
Provenance and Peer Review: This article was commissioned by the Guest Editors (Haotian Lin and Limin Yu) for the series “Medical Artificial Intelligent Research” published in Annals of Translational Medicine. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Deng L, Yu D. Deep learning: methods and applications. Foundations and Trends® in Signal Processing 2014;7:197-387.
- Song HA, Lee SY. Hierarchical representation using NMF. In: International conference on neural information processing. Heidelberg: Springer, 2013:466-73.
- Zhang QL, Zhao D, Chi XB. Review for deep learning based on medical imaging diagnosis. Computer Science 2017;44:1-7.
- Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems. 2012:1097-105.
- Lin M, Chen Q, Yan S. Network in network. arXiv preprint arXiv:1312.4400, 2013.
- Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2015:1-9.
- Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556, 2014.
- Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167, 2015.
- Szegedy C, Vanhoucke V, Ioffe S, et al. Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2016:2818-26.
- Szegedy C, Ioffe S, Vanhoucke V, et al. Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-first AAAI conference on artificial intelligence. 2017.
- He K, Zhang X, Ren S, et al. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans Pattern Anal Mach Intell 2015;37:1904-16. [Crossref] [PubMed]
- He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2016:770-8.
- Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2015:3431-40.
- Everingham M, Van Gool L, Williams CKI, et al. The pascal visual object classes challenge 2012 (voc2012) results. Available online: http://www.pascal-network.org/challenges/VOC/voc2011/workshop/index
- Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention. Cham: Springer, 2015:234-41.
- Badrinarayanan V, Kendall A, Cipolla R. SegNet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 2017;39:2481-95. [Crossref] [PubMed]
- Zhao H, Shi J, Qi X, et al. Pyramid scene parsing network. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2017:2881-90.
- Liskowski P, Krawiec K. Segmenting retinal blood vessels with deep neural networks. IEEE Trans Med Imaging 2016;35:2369-80. [Crossref] [PubMed]
- Fu H, Xu Y, Wong DWK, et al. Retinal vessel segmentation via deep learning network and fully-connected conditional random fields. In: 2016 IEEE 13th international symposium on biomedical imaging (ISBI). New York: IEEE, 2016:698-701.
- Dasgupta A, Singh S. A fully convolutional neural network based structured prediction approach towards the retinal vessel segmentation. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017). New York: IEEE, 2017:248-51.
- Zhu C, Zou B, Zhao R, et al. Retinal vessel segmentation in colour fundus images using extreme learning machine. Comput Med Imaging Graph 2017;55:68-77. [Crossref] [PubMed]
- Hu K, Zhang Z, Niu X, et al. Retinal vessel segmentation of color fundus images using multiscale convolutional neural network with an improved cross-entropy loss function. Neurocomputing 2018;309:179-91. [Crossref]
- Fu H, Cheng J, Xu Y, et al. Joint optic disc and cup segmentation based on multi-label deep network and polar transformation. IEEE Trans Med Imaging 2018;37:1597-605. [Crossref] [PubMed]
- Su Y, Wang Y, Jiao J, et al. Automatic detection and classification of breast tumors in ultrasonic images using texture and morphological features. Open Med Inform J 2011;5:26-37. [Crossref] [PubMed]
- Wang TC, Huang YH, Huang CS, et al. Computer-aided diagnosis of breast DCE-MRI using pharmacokinetic model and 3-D morphology analysis. Magn Reson Imaging 2014;32:197-205. [Crossref] [PubMed]
- Chen SW, Liu YJ, Liu D, et al. AlexNet model and adaptive contrast enhancement based ultrasound imaging classification. Computer Science 2019;46:146-52.
- Shalev-Shwartz S, Zhang T. Stochastic dual coordinate ascent methods for regularized loss minimization. Journal of Machine Learning Research 2013;14:567-99.
- Baumgartner CF, Kamnitsas K, Matthew J, et al. Real-time standard scan plane detection and localisation in fetal ultrasound using fully convolutional neural networks. In: International conference on medical image computing and computer-assisted intervention. Cham: Springer, 2016:203-11.
- Codella NCF, Nguyen QB, Pankanti S, et al. Deep learning ensembles for melanoma recognition in dermoscopy images. IBM Journal of Research and Development 2017;61:5:1-5:15.
- Yang Y, Yan LF, Zhang X, et al. Glioma grading on conventional MR images: a deep learning study with transfer learning. Front Neurosci 2018;12:804. [Crossref] [PubMed]
- Zhang QL, Chi XB, Zhao D. Early diagnosis of Parkinson’s disease based on deep learning. Computer Systems & Applications 2018;27:1-9.
- Sarraf S, Tofighi G. DeepAD: Alzheimer’s disease classification via deep convolutional neural networks using MRI and fMRI. BioRxiv 2016. doi: https://doi.org/. [Crossref]
- Payan A, Montana G. Predicting Alzheimer's disease: a neuroimaging study with 3D convolutional neural networks. arXiv preprint arXiv:1502.02506, 2015.
- Hosseini-Asl E, Ghazal M, Mahmoud A, et al. Alzheimer’s disease diagnostics by a 3D deeply supervised adaptable convolutional network. Front Biosci (Landmark Ed) 2018;23:584-96. [Crossref] [PubMed]
- Poudel RPK, Lamata P, Montana G. Recurrent fully convolutional neural networks for multi-slice MRI cardiac segmentation. In: Zuluaga MA, Bhatia K, Kainz B, et al. Reconstruction, segmentation, and analysis of medical images. Cham: Springer, 2016:83-94.
- Isensee F, Jaeger PF, Full PM, et al. Automatic cardiac disease assessment on cine-MRI via time-series segmentation and domain specific features. In: International workshop on statistical atlases and computational models of the heart. Cham: Springer, 2017:120-9.
- Liao F, Chen X, Hu X, et al. Estimation of the volume of the left ventricle from MRI images using deep neural networks. IEEE Trans Cybern 2019;49:495-504. [Crossref] [PubMed]
- Lieman-Sifry J, Le M, Lau F, et al. FastVentricle: cardiac segmentation with ENet. In: International Conference on Functional Imaging and Modeling of the Heart. Cham: Springer, 2017:127-38.
- Meng Y, Zhang D, Yandong LI, et al. Analysis of ultrasound and pathology images for special types of breast malignant tumors. Chinese Journal of Medical Imaging 2015;(3):188-91.
- Gurcan MN, Boucheron LE, Can A, et al. Histopathological image analysis: a review. IEEE Rev Biomed Eng 2009;2:147-71. [Crossref] [PubMed]
- Zhang Y, Yukun YE, Wang D. Clinical application of image processing and neural network in cytopathological diagnosis of lung cancer. Chinese Journal of Thoracic and Cardiovascular Surgery 2005;04.
- Qaiser T, Mukherjee A, Reddy Pb C, et al. HER2 challenge contest: a detailed assessment of automated HER2 scoring algorithms in whole slide images of breast cancer tissues. Histopathology 2018;72:227-38. [Crossref] [PubMed]
- Gown AM, Goldstein LC, Barry TS, et al. High concordance between immunohistochemistry and fluorescence in situ hybridization testing for HER2 status in breast cancer requires a normalized IHC scoring system. Mod Pathol 2008;21:1271-7. [Crossref] [PubMed]
- Ehteshami Bejnordi B, Balkenhol M, Litjens G, et al. Automated detection of DCIS in whole-slide H&E stained breast histopathology images. IEEE Trans Med Imaging 2016;35:2141-50. [Crossref] [PubMed]
- Yoshida H, Yamashita Y, Shimazu T, et al. Automated histological classification of whole slide images of colorectal biopsy specimens. Oncotarget 2017;8:90719-29. [Crossref] [PubMed]
- Zhang ZZ, Gao JY, Lv G, et al. Pathological image classification of gastric cancer based on depth learning. Computer Science 2018. [Crossref]