
Non-invasive urinary sediment double-immunostaining predicts BK polyomavirus associated-nephropathy in kidney transplant recipients
Introduction
BK polyomavirus associated-nephropathy (BKPyVAN) is one of the main complications affecting renal transplant function and prognosis (1). In kidney transplant recipients, the incidence of BKPyVAN fluctuates between 1% and 10% (2). Based on the Banff classification criteria, BKPyVAN was divided into grade A, grade B and grade C (3). The 2-year renal graft survival is 90%, 70% and 50% for grade A, grade B and grade C respectively (4). Currently, the main screening tests for BK polyomavirus (BKPyV) infection are urine cytology examination, and qualitative and quantitative polymerase chain reaction (PCR) for detecting BKPyV DNA (5). Some centers examine VP1 messenger RNA by real-time PCR (6). Clinical studies showed that the incidence of BKPyV viruria was 20% to 57% (7,8), the incidence of urinary decoy cells was 13% to 42% (9,10), and the incidence of BKPyV viremia was 7% to 29% in kidney transplant recipients (7,11,12). The negative predictive value (NPV) of urine BKPyV DNA for BKPyVAN is nearly 100%, but the positive predictive value (PPV) is only 40% (13). BKPyV viruria usually precedes BKPyV viremia by about 4 weeks, and precedes BKPyVAN by about 12 weeks (13,14). Sustained BKPyV viremia (plasma BKPyV DNA loads >10,000 copies/mL for >4 weeks) had high specificity and sensitivity (both >93%) for BKPyVAN (13,15,16). Urinary polyomavirus-Haufen has been reported to be an accurate indicator for BKPyVAN as it has a 97% PPV and a 100% NPV (17). However, the urinary polyomavirus-Haufen test has not been verified by multiple centers, and requires electron microscopy, which is not routinely available in many centers. As such, kidney biopsy still serves as the golden standard for diagnosing BKPyVAN (5). However, kidney biopsy is an invasive procedure and has limitations including a high risk of bleeding, it can’t be repeated frequently, and has a false negative rate of 36.5% (18).
BKPyV inclusion-bearing epithelial cells, termed “decoy cells”, can be identified on urine cytological examination, using Papanicolaou staining (19). Although, the NPV of urine decoy cells for the diagnosis of BKPyVAN is close to 100%, it’s PPV is only 29% (13). Our previous study observed that a significant number of kidney transplant recipients with positive urinary decoy cells did not have or develop BKPyVAN (20). Therefore, methods of increasing the PPV of urine decoy cells for BKPyVAN is valuable for therapeutic decision.
We speculate that if biomarkers that are specific to renal tubular epithelial cells or urothelial cells could be detected on urinary decoy cells, the source of the decoy cells could be identified. Homogentisate 1, 2-dioxygenase (HGD) is a specific biomarker of tubular epithelial cells (https://www.proteinatlas.org/ENSG00000113924-HGD/tissue) (21), and S100 calcium binding protein P (S100P) is a specific biomarker of urothelial cells (https://www.proteinatlas.org/ENSG00000163993-S100P/tissue) (22-24). In voided urinary sediment samples from patients with biopsy-proven BKPyVAN, we observed HGD(+)/SV40-T(+) cells and S100P(+)/SV40-T(+) cells by double-immunostaining. It can be reasonably considered that HGD(+)/SV40-T(+) cells represent BKPyV-infected tubular epithelial cells, and S100P(+)/SV40-T(+) cells represent BKPyV-infected urinary transitional epithelial cells.
We hypothesize that the presence of urinary HGD(+)/SV40-T(+) cells indicate an active intra-renal BKPyV infection. This study was performed to identify the source of urinary decoy cells by double-immunostaining using anti-S100P or anti-HGD, and anti-SV40-T antibodies. Furthermore, we sought to determine if the presence of urinary HGD(+)/SV40-T(+) cells were predictive of BKPyVAN.
Methods
Study population
This retrospective cross-sectional study was performed in a single center. The inclusion criteria included: a history of kidney transplantation, urinary decoy cells ≥1/10 HPF (high power field), and urinary BKPyV DNA load >103 copies/mL. The exclusion criteria included: recurrent BKPyVAN, repeated kidney transplant, and lack of renal biopsy. Isolation of urinary sediment cells was performed within 3 days before/after renal biopsy. This study was approved by the Ethics Committee and the Research Board of our institution.
Urine cytology
Papanicolaou staining method was used for urinary cytological smears. Decoy cells were counted as the number per 10 HPFs (25). The criteria for the diagnosis of decoy cells was an obviously enlarged and malformed nucleus of BKPyV-infected cells, with the presence of basophilic viral inclusions.
Virological studies
Urine and plasma BKPyV load were quantitatively measured by quantitative PCR (MJ Research, Waltham, MA, USA). Specimen collection and processing, PCR primers, TaqMan probe (targeting the BKPyV VP1 gene), plasmid standard containing the targeted BKPyV VP1 gene, amplification protocols, PCR precautions, and quality assurance were performed as previously described (26). Urine or plasma BKPyV load were presented as BKPyV genome copies per milliliter. The lower limit of quantitation was determined at 1,000 copies per milliliter.
Pathological diagnosis of BKPyVAN
The pathological diagnosis of BKPyVAN was confirmed by immunohistochemical (IHC) staining with anti-SV40-T large T antigen monoclonal antibody, as previously described (27). The histological features of BKPyVAN were classified using the American Society of Transplantation (AST) schema; BKPyVAN was classified as A, B, and C based on the guidelines published by Hirsch et al. (28).
Antibodies
Using the database of the Human Protein Atlas (http://www.proteinatlas.org/) (29), we screened more than 40 biomarkers that are mainly expressed in either transitional epithelium or in renal tubular epithelium. Based on our review, we selected S100P as a specific marker of transitional epithelial cells (https://www.proteinatlas.org/ENSG00000163993-S100P/tissue) (22-24), and HGD as a specific marker of renal tubular epithelial cells (https://www.proteinatlas.org/ENSG00000113924-HGD/tissue) (21). BKPyV was detected by anti-SV40-T large antigen antibody. The results of double-immunostaining with anti-S100P antibody + anti-SV40-T antibody or anti-HGD antibody + anti-SV40-T antibody on ureterocystic and renal tissue with BKPyVAN and without BKPyVAN were presented in Figure S1 and Figure S2.
Preparation of cell block
Patients were instructed to collect a total of 600 mL of fresh urine in a sterile bottle from 10:00 pm to 8:00 am. To prevent cell lysis, 60 mL of 4% formaldehyde solution was added to the urine specimen. The urine specimen was centrifuged for 10 min at 500 ×g within 2 h after collection. The urine sediment was transferred to EP tube with 2 mL of 4% formaldehyde solution. After centrifuging (at 500 ×g for 2 min), the supernatant was discarded.
The Anbiping kit (LBP Medicine Science & Technology Co., Ltd., Guangzhou, China, Batch number 1708001) was used for cell block preparation. The procedure was performed strictly in accordance with the instructions. A total of 0.15 mL of Reagent A was added to the urine sediment. The mixture was shaken and centrifuged for 2 min at 500 ×g. After discarding the supernatant, 0.15 mL of Reagent B was added to the urine sediment. After standing for 1 min, the cell mass was transferred to a cassette for dehydration (Leica ASP300S). The detailed procedure of dehydration was as follows.
- Neutral formalin, 10 min,
- 70% ethyl alcohol, 20 min,
- 80% ethyl alcohol, 20 min,
- 90% ethyl alcohol I, 20 min,
- 90% ethyl alcohol II, 20 min,
- 100% ethyl alcohol I, 20 min,
- 100% ethyl alcohol II, 20 min,
- Xylene I, 20 min,
- Xylene II, 20 min,
- Paraffin I, 30 min,
- Paraffin II, 30 min,
- Paraffin III, 30 min.
Paraffin-embedded section was sliced at 3 µm (Leica, RM2235) and then was dried at 65 °C for 30 min (Leica, HI1220).
Double immunohistochemical staining
Double-immunostaining was performed using a fully automatic IHC staining machine (Leica Band-Max, Germany). The automatic double staining protocol (Sequential double staining) was set as follows.
Part 1 Equivalent to the following operating procedure step V) to step XIX) (Bond Polymer Refine IHC protocol, Leica, Microsystems, batch number 62798)
- Marker 1 (anti-SV40-T mouse monoclonal antibody, 1:1,000, Catalogue Number DP02, Merck Millipore, USA) 15 min,
- Post Primary, 8 min,
- Polymer, 8 min,
- Peroxide Block, 5 min,
- Mixed DAB Refine, 5 s,
- Mixed DAB Refine, 5 min.
Part 2 Equivalent to the following operating procedure step XXII) to step XXVI) (Bond Polymer Refine Red IHC protocol, Leica, Microsystems, batch number 65131)
- Marker 2 (anti-S100P rabbit monoclonal antibody, 1:5,000, Catalogue Number ab133554, Abcam, Cambridge, UK; or anti-HGD rabbit monoclonal antibody, 1:3,200, Catalogue Number ab131035, Abcam, Cambridge, UK) 15 min,
- Post Primary alkaline phosphatase (AP) 20 min,
- Polymer AP 30 min,
- Mixed Red Refine 10 min,
- Mixed Red Refine 5 min,
- Hematoxylin 6 min.
The detailed procedure of double-immunostaining (anti-SV40-T + anti-S-100P or anti-SV40-T + anti-HGD) was as follows
- Deparaffinizing with Bond solution,
- Rinsing with 100% alcohol for 1 min, 3 times,
- Rinsing with Bond wash solution for 1 min, 3 times,
- Antigen retrieval with Bond epitope retrieval solution for 20 min (pH 6.0, 100 °C), followed by washing with wash buffer for 1 min, 3 times,
- Incubating with anti-SV40-T antibody, 15 min (37 °C),
- Rinsing with Bond wash solution for 1 min, 3 times,
- Post-primary, 8 min,
- Rinsing with Bond wash solution for 1 min, 3 times,
- Polymer, 8 min,
- Rinsing with Bond wash solution for 1 min, 3 times,
- Peroxide block, 5 min,
- Rinsing with Bond wash solution for 1 min, 3 times,
- Mixed DAB Refine, 5 s,
- Mixed DAB Refine, 5 min,
- Rinsing with Bond deionized water solution for 1 min, 3 times,
- Rinsing with Bond wash solution for 1 min, 3 times,
- Incubating with the anti-S100P antibody or anti-HGD, 15 min (37 °C),
- Rinsing with Bond wash solution for 1 min, 3 times,
- Post Primary AP, 20 min,
- Rinsing with Bond wash solution for 1 min, 3 times,
- Post Primary AP, 20 min,
- Rinsing with Bond wash solution for 1 min, 3 times,
- Polymer AP, 30 min,
- Rinsing with Bond wash solution for 1 min, 3 times,
- Mixed Red Refine, 10 min,
- Mixed Red Refine, 5 min,
- Rinsing with Bond deionized water solution for 1 min, 3 times,
- Rinsing with Bond wash solution for 1 min, 3 times,
- Hematoxylin, 6 min
- Rinsing with Bond deionized water solution for 1 min, 3 times,
- Air drying,
- Transparentizing with xylene,
- Mounted with resin.
IHC slides were reviewed by 2 independent cytopathologists who were blinded to the histological diagnosis and clinical data.
Statistical analysis
Clinical data was expressed as count (percentage), or as mean ± standard deviation. Sensitivity, specificity, PPV, NPV, and 95% confidence intervals (CIs) of urinary HGD(+)/SV40-T(+) cells for diagnosing BKPyVAN were calculated using renal allograft biopsy findings as the gold standard. Correlation analysis between HGD(+)/SV40-T(+) cells and various pathological damage indicators was evaluated by a Chi-square test. IBM SPSS version 20 software (IBM Corporation, Somers, New York, USA) was used for statistical analyses. Values of P<0.05 were considered statistically significant.
Results
Patient characteristics
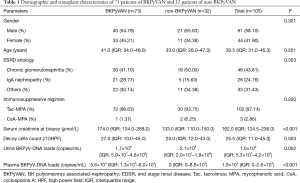
Demographic and transplant characteristics are shown in Table 1. A total of 105 urine samples from 105 patients [44 females, 61 males; median age 40 years, interquartile range (IQR): 31–46 years] were studied morphologically by double-immunostaining. The median time of detecting positive decoy cells, namely patients’ enrollment, was 10.9 months (IQR: 5.2–24.6 months) after kidney transplantation. According to the pathological results, all patients were divided into 73 cases of BKPyVAN and 32 cases of non-BKPyVAN. The BKPyVAN group consists of 5 cases of stage A, 60 cases of stage B, and 8 cases of stage C. The median initial serum creatinine level was 162.0 (125.3–235.0) µmol/L, the number of decoy cells was 23/10 HPF (IQR: 11–39), and the BKPyV viral load in the urine was 1.0×109 copies/mL (IQR: 5.3×107–4.2×109) and in the plasma was 1.9×103 copies/mL (IQR: 0–2.6×104).
Full table
Urinary decoy cells
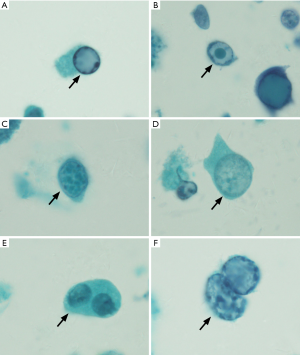
On Papanicolaou staining, the decoy cells exhibited 4 classical morphological types (30). Type 1 classic decoy cell contained enlarged, basophilic, homogeneous amorphous ground glass-like intra-nuclear inclusion body (Figure 1A). Type 2 decoy cell contained a single nuclear inclusion body surrounded by a peripheral halo, appearing as a bird’s eye showing (Figure 1B). Type 3 decoy cell contained scattered clumped chromatin (Figure 1C). Type 4 are decoy cell contained cytoplasmic vesicles with fine-grained chromatin and nucleoli (Figure 1D).
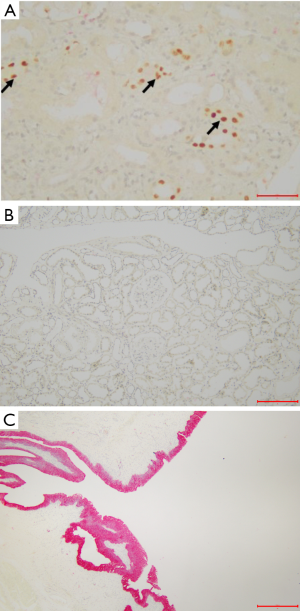
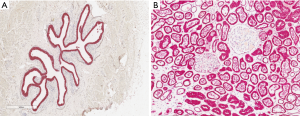
S100P(+) cells and HGD(+) cells in renal tissue without BKPyVAN
To confirm the specificity of S100P for urothelial epithelium and HGD for tubular epithelium, tissue sections from normal bladder, ureter, and kidney were examined. Positive anti-S100P immunostaining was detected in the cytoplasm of urothelial cells (red) (Figure 2A). Inversely, positive anti-HGD immunostaining was detected in the cytoplasm of tubular epithelial cells (red) (Figure 2B).
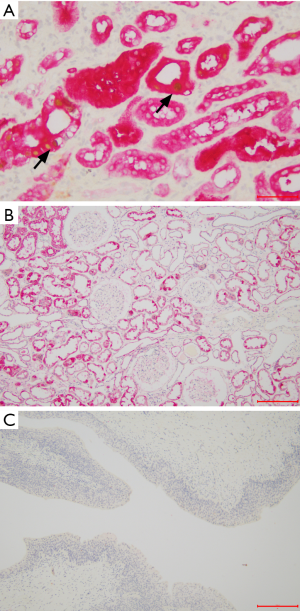
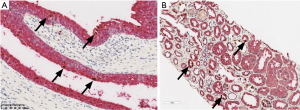
S100P(+)/SV40-T(+) cells and HGD(+)/SV40-T(+) cells in renal tissue with BKPyVAN
The tissue sections from BKPyV-infected kidneys, ureter, and bladder were examined to confirm the co-expression of HGD/SV40-T in tubular epithelial cells and S100P/SV40-T in urothelial cells. In BKPyV-infected urothelial cells, the cytoplasm was positive for S100P (red) and the nuclei were positive for SV40-T (brown) (Figure 3A). On the other hand, in BKPyV-infected kidney samples the cytoplasm was positive for HGD (red) and the nuclei were positive for SV40-T (brown) (Figure 3B).
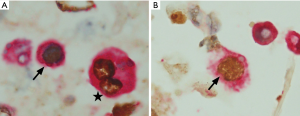
S100P(+)/SV40-T(+) cells and HGD(+)/SV40-T(+) cells in cell block
Typical decoy cells detected by double-immunostaining are shown in Figure 4. Double-immunostaining results were qualitatively reported as either present (one or more cells) or absent. In urine samples, the BKPyV-infected (SV40-T-positive) cells were categorized as S100P(+)/SV40-T(+) (Figure 4A) and HGD(+)/SV40-T(+) (Figure 4B), and HGD(−)/S100P(-)/SV40-T(+). Of the 105 patients, 76 (72.4%) patients had HGD(+)/SV40-T(+) cells and S100P(+)/SV40-T(+) cells simultaneously (group A), 24 (22.9%) patients had S100P(+)/SV40-T(+) cells only (group B), and 5/109 (4.6%) patients had S100P(−)/HGD(−)/SV40-T(+) cells only (group C). All patients with HGD(+)/SV40-T(+) cells also had S100P(+)/SV40-T(+) cells. Conversely, among patients with S100P(+)/SV40-T(+) cells, only 76 (76%) patients also had HGD(+)/SV40-T(+) cells. In most cases, the proportion of S100P(+)/SV40-T(+) cells was significantly higher than that of HGD(+)/SV40-T(+) cells.
Predictive value of urinary HGD(+)/SV40-T(+) cells for the diagnosis of BKPyVAN
In group A, 70 (92.1%) patients were diagnosed with BKPyVAN. In group B, 3 (12.5%) patients were diagnosed with BKPyVAN. In group C, no patient was diagnosed with BKPyVAN. The PPV, NPV, sensitivity, and specificity of urinary HGD(+)/SV40-T(+) cells for predicting BKPyVAN were 92.1% (95% CI: 83.0–96.7%), 89.7% (95% CI: 71.5–97.3%), 95.9% (95% CI: 87.7–98.9%), and 81.3% (95% CI: 63.0–92.1%).
Correlation analysis between HGD(+)/SV40-T(+) cells and various pathological damage indicators showed that HGD(+)/SV40-T(+) cells were correlated with the extent of SV40-T (P<0.001) and tubulitis (P=0.001), but not with interstitial inflammation (P=0.734), total interstitial inflammation (P=0.739), tubular atrophy (P=0.366), or interstitial fibrosis (P=0.505).
Diagnostic performance of urinary decoy cells and BKPyV viremia for the diagnosis of BKPyVAN
The area under the ROC curve of predicting BKPyVAN by decoy cells was 0.531 (0.431–0.630), with an optimal cut-off value of 29 (per 10 high power field), a sensitivity of 45.8% (95% CI: 34.0–58.0%), and a specificity of 68.8% (95% CI: 50.0–83.9%). Besides, the area under the ROC curve of predicting BKPyVAN by plasma BKPyV loads was 0.735 (95% CI: 0.632–0.822), with an optimal cut-off value of 1000 copies/mL, a sensitivity of 61.1% (95% CI: 48.9–72.4%) and a specificity of 84.2% (95% CI: 60.4–96.6%).
Discussion
The detection of decoy cells in urine is generally used as a simple and inexpensive technique for screening BKPyV viruria after kidney transplantation. However, Hirsch et al. reported that the PPV of a “positive” decoy cells for predicting BKPyVAN was 25% to 30% (13). Decoy cells may be derived from urothelium and/or the transplanted kidney (31), and it is difficult to identify the source of decoy cells by morphological appearance alone. Double-immunostaining with anti-P16 and anti-Ki67 antibodies is widely used for screening and diagnosing cervical precancerous lesions and cervical cancer (32). Inspired by this idea, we put forward a hypothesis that detecting decoy cells with double-immunostaining targeting tubule-specific or urothelium-specific markers can identify the source of the decoy cells.
S100P is a member of the S100 calcium-binding protein P family, and functions by regulating cell growth and differentiation (33). In the urinary system, S100P mainly expresses in renal pelvic urothelium, ureteral epithelium, and bladder urothelium, but not in tubular epithelium or glomerular parietal epithelial cells (https://www.proteinatlas.org/ENSG00000163993-S100P/tissue). HGD is a member of the 1 ensembl protein family, and functions by regulating the catabolism of the amino acids tyrosine and phenylalanine (34). In the urinary system, HGD is mainly expressed in the cytoplasm of proximal and distal tubular, and collecting duct epithelium, but is not expressed in renal pelvic urothelium, ureteral epithelium, and bladder urothelium cells (https://www.proteinatlas.org/ENSG00000113924-HGD/tissue). In our study, IHC staining results confirmed that HGD was only expressed in renal tubular epithelial cells, and S100P was only expressed in ureter and bladder epithelial cells. Therefore, it is reasonable to believe that HGD and S100p are useful markers for differentiating renal tubular epithelial cells from urothelial cells, and that the presence of HGD antigen in the cytoplasm of decoy cells strongly indicates that these cells are detached tubular epithelial cells.
Our results showed that the qualitative detection of HGD(+)/SV40-T(+) cells in a fixed urine cell block predicted BKPyVAN with high PPV (92.1%) and high NPV (89.7%). Actually, the PPV may even be higher because of the local distribution of BKPyV replication in renal allograft and potential sample error. In this study, 3 patients had positive urinary HGD(+)/SV40-T(+) cells, but negative anti-SV40-T IHC staining in renal allograft biopsy specimens, which we believe to be misdiagnosed BKPyVAN because of sample error.
Some researchers believe that BKPyV remains latent in tubular epithelial cells and urothelial cells. Once host immunity is inhibited, the latent BKPyV reactivates and replicates, resulting in the host cells dropping into the urine to form decoy cells (35). Others believe that decoy cells mainly originate from bladder epithelial cells and travels in a retrograde manner along the ureter to infect tubular epithelial cells (36). In our study, HGD(+)/SV40-T(+) and S100P(+)/SV40-T(+) double-immunostaining was performed on urine sediment respectively, and the results showed that the proportion of S100P (+)/SV40-T (+) cells was greater than that of HGD(+)/SV40-T(+) cells. In addition, S100P(+)/SV40-T(+) cells were found in all patients with HGD(+)/SV40-T(+) cells. However, in 100 patients with S100P(+)/SV40-T(+) cells, HGD(+)/SV40-T(+) cells were not found in 24 patients. We believe that this group of patients have an early BKPyV-activated infection, only involving the bladder and/or ureteral transitional epithelial cells. As such, our results demonstrated that in most patients with urinary BKPyV infection, decoy cells are predominantly derived from the urothelium. This phenomenon appears to be inconsistent with the results of a previous study in which only anti-S100P and anti-SV40-T antibodies, but no antibody aimed at renal tubular epithelial cells, were used for detecting decoy cells (37). In that study, the authors examined urine samples from 7 kidney transplant recipients for S100P(+)/SV40-T(+) cells and S100P(−)/SV40-T(+) cells, and the S100P(+)/SV40-T(+) cells accounted for a relatively low proportion of all SV40-T(+) cells, so they indirectly inferred that S100P(−)/SV40-T(+) cells are derived from the renal tubular epithelium and most decoy cells are tubular-derived cells.
Notably, in a few cases, BKPyV can infect the glomeruli. Both Celik et al. and Ravindran et al. found that BKPyV could involve glomeruli in biopsy of transplanted kidney (38,39). In addition, Alcendor et al. and Popik et al. also demonstrated that BKPyV could infect and replicate in glomerular cells in vitro (40,41). Similarly, in our previous study, we also found that BKPyV could infect glomerular parietal epithelial cells, and this phenomenon often indicated more serious pathological damage and worse prognosis of transplanted kidney function (42). Moreover, our results showed that BKPyV infection in renal tubular epithelial cell is concurrent in all patients with glomerular BKPyV involvement. This may be because BKPyV usually spreads in a retrograde way. Theoretically, the combination of HGD and SV40-T is enough to demonstrate whether BKPyV replicate in transplanted kidney or not. Of course, we also expect to find a specific marker of glomerulus, which can provide us with clues of glomerular BKPyV infection.
The novel, non-invasive urine-based BKPyV-Haufen test has been reported to diagnose BKPyVAN with a PPV and NPV both greater than 95% (17). BKPyV-Haufen form in virus-infected renal tubules are flushed out of the kidney, and appear in the urine as a direct marker of intra-renal viral disease (17,43). Similar to the detection of BKPyV-Haufen aggregates, urinary HGD(+)/SV40-T(+) cells detach from virally injured tubular epithelium and subsequently are flushed out of the kidney. Thus, the presence of urinary HGD(+)/SV40-T(+) cells directly reflect BKPyV replication in renal allografts. Detection of urinary polyomavirus-Haufen requires an experienced electron microscopist, and electron microscopy can’t be routinely performed in many transplant centers. In contrast, double-immunostaining is simple to perform, can be done at almost every transplant center, and even an inexperienced pathologist can make a diagnosis. The technique can be used as a screening tool for BKPyVAN during outpatient follow-up, and as a supplementary test when a patient is suspected to have BKPyVAN but biopsy samples are negative.
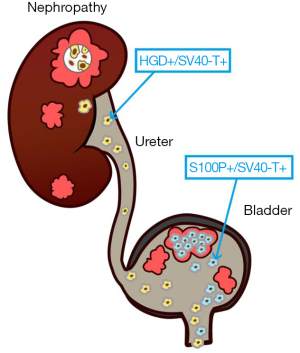
In conclusion, urinary decoy cell identification via double-immunostaining represents a reliable alternative method for determining the origin of decoy cells in urine samples and for predicting BKPyVAN (Figure 5). We believe that this non-invasive method combined with PCR can provide useful information for diagnosing BKPyVAN when renal biopsy specimens are IHC staining negative for anti-SV40-T. Moreover, regular monitoring of urine HGD(+)/SV40-T(+) cells after transplantation cells can further demonstrate its diagnostic performance for BKPyVAN.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (81770749), the Natural Science Foundation of Guangdong Province (2017A030313710), the Basic Scientific Research Fund of Sun Yat-Sen University (17ykpy29) and the China Organ Transplantation Development Foundation (2019JYJH07).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are responsible for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee and the Research Board, the First Affiliated Hospital of Sun Yat-sen University {No. [2019]220}.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yi SG, Knight RJ, Lunsford KE. BK virus as a mediator of graft dysfunction following kidney transplantation. Curr Opin Organ Transplant 2017;22:320-7. [Crossref] [PubMed]
- Hirsch HH, Brennan DC, Drachenberg CB, et al. Polyomavirus-associated nephropathy in renal transplantation: interdisciplinary analyses and recommendations. Transplantation 2005;79:1277-86. [Crossref] [PubMed]
- Sar A, Worawichawong S, Benediktsson H, et al. Interobserver agreement for Polyomavirus nephropathy grading in renal allografts using the working proposal from the 10th Banff Conference on Allograft Pathology. Hum Pathol 2011;42:2018-24. [Crossref] [PubMed]
- Nickeleit V, Singh HK. Polyomaviruses and disease: is there more to know than viremia and viruria? Curr Opin Organ Transplant 2015;20:348-58. [Crossref] [PubMed]
- Sawinski D, Goral S. BK virus infection: an update on diagnosis and treatment. Nephrol Dial Transplant 2015;30:209-17. [Crossref] [PubMed]
- Bateman AC, Greninger AL, Atienza EE, et al. Quantification of BK Virus Standards by Quantitative Real-Time PCR and Droplet Digital PCR Is Confounded by Multiple Virus Populations in the WHO BKV International Standard. Clin Chem 2017;63:761-9. [Crossref] [PubMed]
- Bressollette-Bodin C, Coste-Burel M, Hourmant M, et al. A prospective longitudinal study of BK virus infection in 104 renal transplant recipients. Am J Transplant 2005;5:1926-33. [Crossref] [PubMed]
- Herman J, Van Ranst M, Snoeck R, et al. Polyomavirus infection in pediatric renal transplant recipients: evaluation using a quantitative real-time PCR technique. Pediatr Transplant 2004;8:485-492. [Crossref] [PubMed]
- Viscount HB, Eid AJ, Espy MJ, et al. Polyomavirus polymerase chain reaction as a surrogate marker of polyomavirus-associated nephropathy. Transplantation 2007;84:340-5. [Crossref] [PubMed]
- Huang G, Chen LZ, Qiu J, et al. Prospective study of polyomavirus BK replication and nephropathy in renal transplant recipients in China: a single-center analysis of incidence, reduction in immunosuppression and clinical course. Clin Transplant 2010;24:599-609. [Crossref] [PubMed]
- Geddes CC, Gunson R, Mazonakis E, et al. BK viremia surveillance after kidney transplant: single-center experience during a change from cyclosporine-to lower-dose tacrolimus-based primary immunosuppression regimen. Transpl Infect Dis 2011;13:109-16. [Crossref] [PubMed]
- Babel N, Fendt J, Karaivanov S, et al. Sustained BK viruria as an early marker for the development of BKV-associated nephropathy: analysis of 4128 urine and serum samples. Transplantation 2009;88:89-95. [Crossref] [PubMed]
- Hirsch HH, Knowles W, Dickenmann M, et al. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N Engl J Med 2002;347:488-96. [Crossref] [PubMed]
- Brennan DC, Agha I, Bohl DL, et al. Incidence of BK with tacrolimus versus cyclosporine and impact of preemptive immunosuppression reduction. Am J Transplant 2005;5:582-94. [Crossref] [PubMed]
- Randhawa PS, Finkelstein S, Scantlebury V, et al. Human polyoma virus-associated interstitial nephritis in the allograft kidney. Transplantation 1999;67:103-9. [Crossref] [PubMed]
- Hirsch HH, Steiger J, Polyomavirus BK. Lancet Infect Dis 2003;3:611-23. [Crossref] [PubMed]
- Singh HK, Andreoni KA, Madden V, et al. Presence of urinary Haufen accurately predicts polyomavirus nephropathy. J Am Soc Nephrol 2009;20:416-27. [Crossref] [PubMed]
- Drachenberg CB, Papadimitriou JC, Hirsch HH, et al. Histological patterns of polyomavirus nephropathy: correlation with graft outcome and viral load. Am J Transplant 2004;4:2082-92. [Crossref] [PubMed]
- Becker GJ, Garigali G, Fogazzi GB. Advances in Urine Microscopy. Am J Kidney Dis 2016;67:954-64. [Crossref] [PubMed]
- Huang G, Chen WF, Wang CX, et al. Noninvasive tool for the diagnosis of polyomavirus BK-associated nephropathy in renal transplant recipients. Diagn Microbiol Infect Dis 2013;75:292-7. [Crossref] [PubMed]
- Fagerberg L, Hallstrom BM, Oksvold P, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 2014;13:397-406. [Crossref] [PubMed]
- Higgins JP, Kaygusuz G, Wang L, et al. Placental S100 (S100P) and GATA3: markers for transitional epithelium and urothelial carcinoma discovered by complementary DNA microarray. Am J Surg Pathol 2007;31:673-80. [Crossref] [PubMed]
- Chuang AY, DeMarzo AM, Veltri RW, et al. Immunohistochemical differentiation of high-grade prostate carcinoma from urothelial carcinoma. Am J Surg Pathol 2007;31:1246-55. [Crossref] [PubMed]
- Gulmann C, Paner GP, Parakh RS, et al. Immunohistochemical profile to distinguish urothelial from squamous differentiation in carcinomas of urothelial tract. Hum Pathol 2013;44:164-72. [Crossref] [PubMed]
- Nickeleit V, Klimkait T, Binet IF, et al. Testing for polyomavirus type BK DNA in plasma to identify renal-allograft recipients with viral nephropathy. N Engl J Med 2000;342:1309-15. [Crossref] [PubMed]
- Huang G, Wang CX, Zhang L, et al. Monitoring of polyomavirus BK replication and impact of preemptive immunosuppression reduction in renal-transplant recipients in China: a 5-year single-center analysis. Diagn Microbiol Infect Dis 2015;81:21-6. [Crossref] [PubMed]
- Huang G, Wu LW, Yang SC, et al. Factors Influencing Graft Outcomes Following Diagnosis of Polyomavirus -Associated Nephropathy after Renal Transplantation. PLoS One 2015;10:e0142460. [Crossref] [PubMed]
- Hirsch HH, Randhawa P. BK polyomavirus in solid organ transplantation. Am J Transplant 2013;13 Suppl 4:179-88. [Crossref] [PubMed]
- Pontén F, Jirstrom K, Uhlen M. The Human Protein Atlas--a tool for pathology. J Pathol 2008;216:387-93. [Crossref] [PubMed]
- Singh HK, Bubendorf L, Mihatsch MJ, et al. Urine cytology findings of polyomavirus infections. Adv Exp Med Biol 2006;577:201-12. [Crossref] [PubMed]
- Lamarche C, Orio J, Collette S, et al. BK Polyomavirus and the Transplanted Kidney: Immunopathology and Therapeutic Approaches. Transplantation 2016;100:2276-87. [Crossref] [PubMed]
- Walts AE, Lechago J, Bose S. P16 and Ki67 immunostaining is a useful adjunct in the assessment of biopsies for HPV-associated anal intraepithelial neoplasia. Am J Surg Pathol 2006;30:795-801. [Crossref] [PubMed]
- Suryavanshi M, Sanz-Ortega J, Sirohi D, et al. S100P as a Marker for Urothelial Histogenesis: A Critical Review and Comparison With Novel and Traditional Urothelial Immunohistochemical Markers. Adv Anat Pathol 2017;24:151-60. [Crossref] [PubMed]
- Nemethova M, Radvanszky J, Kadasi L, et al. Twelve novel HGD gene variants identified in 99 alkaptonuria patients: focus on 'black bone disease' in Italy. Eur J Hum Genet 2016;24:66-72. [Crossref] [PubMed]
- Doerries K. Human polyomavirus JC and BK persistent infection. Adv Exp Med Biol 2006;577:102-16. [Crossref] [PubMed]
- Herawi M, Parwani AV, Chan T, et al. Polyoma virus-associated cellular changes in the urine and bladder biopsy samples: a cytohistologic correlation. Am J Surg Pathol 2006;30:345-50. [Crossref] [PubMed]
- Ariyasu S, Yanai H, Sato M, et al. Simultaneous immunostaining with anti-S100P and anti-SV40 antibodies revealed the origin of BK virus-infected decoy cells in voided urine samples. Cytopathology 2015;26:250-5. [Crossref] [PubMed]
- Celik B, Randhawa PS. Glomerular changes in BK virus nephropathy. Hum Pathol 2004;35:367-70. [Crossref] [PubMed]
- Ravindran A, Kashtan CE, Sethi S. BK nephropathy with glomerular involvement. Kidney Int 2018;94:432. [Crossref] [PubMed]
- Alcendor DJ. BK Polyomavirus Virus Glomerular Tropism: Implications for Virus Reactivation from Latency and Amplification during Immunosuppression. J Clin Med 2019;8. [Crossref] [PubMed]
- Popik W, Khatua AK, Fabre NF, et al. BK Virus Replication in the Glomerular Vascular Unit: Implications for BK Virus Associated Nephropathy. Viruses 2019;11. [Crossref] [PubMed]
- Chen XT, Yang SC, Chen WF, et al. Glomerular Parietal Epithelial Cells Infection Is Associated With Poor Graft Outcome in Kidney Transplant Recipients With BK Polyomavirus-Associated Nephropathy. J Infect Dis 2019;219:1879-86. [Crossref] [PubMed]
- Singh HK, Reisner H, Derebail VK, et al. Polyomavirus nephropathy: quantitative urinary polyomavirus-Haufen testing accurately predicts the degree of intrarenal viral disease. Transplantation 2015;99:609-15. [Crossref] [PubMed]


