
Building radiation-resistant model in triple-negative breast cancer to screen radioresistance-related molecular markers
Introduction
Breast cancer is the most common malignancy in women, accounting for about 25%; also, it is the leading cause of tumor deaths among women (1). Radiotherapy plays key roles in the multidisciplinary treatment of breast cancer and mainly reduces the mortality of breast cancer patients by decreasing local recurrence rate and distant metastasis rate (2,3).
Several studies have shown that triple-negative breast cancer (TNBC) is highly invasive, and its distant metastasis rate and local recurrence rate are significantly higher than those of luminal-type breast cancer, along with worse prognosis than luminal-type breast cancer (4-9). Our studies have demonstrated that the radiosensitivity of breast cancer is related to its molecular type, HER2 overexpression and TNBC are independent risk factors for poor prognosis (7,8). A Danish study also obtained similar result: the authors found luminal-type breast cancer patients were more likely to benefit from postoperative radiotherapy than those with TNBC (10). In our radiobiological researches, the radiosensitivity of the TNBC cell line was worse than that of luminal-type breast cancer cell line (11). All of these results have suggested that, compared with luminal-type breast cancer, there may be some degree of radioresistance in TNBC, and such radioresistance may be one of the most important reasons that restrict the therapeutic efficacy on TNBC.
Therefore, it is imperious to explore the possible mechanism of TNBC radiosensitivity of malignant tumors by establishing effective radiobiological models in vitro and in vivo. Several literatures have described the establishment of radioresistance models, and the modeling methods often differed among different studies (12,13). In most studies, the parent cell lines were exposed to a single high-dose irradiation or were repeatedly induced by a certain dose of radiation, and eventually the radioresistant cell lines were obtained. In our previous researches, we had repeatedly irradiated MDA-MB-231 with a certain dose of high-energy rays to obtain radioresistant TNBC cell subtypes, but that could not be stably passed. In our current study, we would further explore the method of maintaining the stability of radioresistance in the radioresistant cell line and constructed a stable cell and mice model for the subsequent experiments.
We compared the differentially expressed genes between MDA-MB-231 cell (231 cell) and radioresistant MDA-MB-231 cell subtype (231-RR cell), and tried to investigate the possible mechanism of radioresistance of TNBC. In addition, we used a gemcitabine-resistant TNBC MDA-MB-231 cell subtype (231-GEM cell) (14). The parental 231, radioresistant 231-RR, and gemcitabine-resistant 231-GEM cells were simultaneously detected with array-based gene expression profiling to screen for differentially expressed genes in three TNBC subtype lines. Meanwhile, we tried to apply a novel radioresistant xenograft induction mode in nude mice and obtained radioresistant breast cancer xenograft, tumor tissue which was used for verifying the differential genes obtained during the prophase screening.
Generally, breast cancer patients with radioresistance or high expressions of chemoresistance-related genes tend to have poor prognosis, whereas the prognosis is relatively good in patients with high expressions of radiosensitivity- or chemosensitivity-associated genes (15,16). Therefore, we verified our findings with the clinical breast cancer specimens in The Cancer Genome Atlas (TCGA), with an attempt to determine the relationship between the screened radioresistance-related genes and the prognosis of breast cancer patients and search for markers that can be used for predicting the radiosensitivity of breast cancer patients and for potential radiosensitization targets.
Methods
Cell lines and cell culture
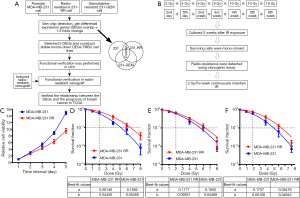
Human TNBC cell line MDA-MB-231 (ATCC® HTB-26™) was purchased from America Type Culture Collection (ATCC). Gemcitabine-resistant TNBC strain MDA-MB-231-GEM was constructed and preserved by Fudan University Breast Cancer Institute (14). The radioresistant TNBC cell strain MDA-MB-231-RR was constructed by our research team. The MDA-MB-231 cells were cultured in GIBCO® RPMI Media 1640 containing 10% fetal bovine serum (FBS) (GIBCO, Invitrogen), 100 U/mL penicillin, and 100 mg/mL streptomycin. The cells were placed in 37 °C in an environment with saturated humidity and 5% CO2. Cells adhered to the wall of culture dish. The design and flow chart of this research was shown in Figure 1A.
Irradiation methods of cells and nude mice
Cells and mice were irradiated by using a special platform (Small animal radiation research platform, SARRP, Gulmay Medical Co., Ltd), with the following parameters: 3.845 Gy/min at 220 kV, 13 mA; the source-to-skin distance (SSD) for a fixed fluoroscopy unit was 35 cm, and the irradiation area was 15 cm × 15 cm. The nude mice were anesthetized by using 1% pentobarbital sodium (0.1 mL/20 g) before irradiation. The xenograft tumor was fully exposed in the irradiation field. All the irradiation procedures were performed at room temperature.
Construction of radioresistant TNBC cell line MDA-MB-231-RR
MDA-MB-231 cells received intermittent irradiation at a single dose of 2 → 4 → 6 → 8 → 10 → 10 → 10 Gy for 6 times, and the total irradiation dose reached 50 Gy. The 2 → 4 Gy irradiation was completed in the 1st week, with a 2-day interval. The irradiation doses 6 → 8 → 10 → 10 → 10 Gy were applied weekly, and the culture continued for 1–2 weeks after the last irradiation. The surviving cells were mono-cloned and named MDA-MB-231-RR. The cells were passed continuously under low-dose radiation (2 Gy/week). The radiosensitivity of MDA-MB-231-RR was detected by clonogenic and cell proliferation assay. If the radioresistance of MDA-MB-231-RR disappeared or weakened, the surviving cells received supplementary irradiation at same dose series until the remarkably radioresistant 231-RR cells were obtained. The flow chart was shown in Figure 1B.
Clonogenic assay
MDA-MB-231 cells received the irradiation (0, 1, 2, 4, 6, and 8 Gy), they were inoculated in an incubator for 12–15 days. After washing with aseptic PBS 1–2 times, 0.5‰ methyl blue dye solution was added before staining for 15–30 min. Then, the dye was removed, and the cells were dried at room temperature. Clones containing ≥50 cells were counted, and the survival fraction of cells was calculated. The plating efficiency (PE) = number of colonies formed when not irradiated/number of inoculating cells × 100%. Then, the survival fraction (SF) = number of colonies at a specific dose/(number of inoculating cells × PE). The linear-quadratic model (L-Q model) was used to fit the curve by using the following model fitting equation: y=exp[−(α*X+β*X2)].
Measurement of cell proliferation
The cell suspensions were inoculated into a 96-well plate, with 100 µL in each well. Cells were continuously cultured in a 37 °C and 5% CO2 incubator. In another blank control group, no cell was incubated and only 100 µL of medium was used as the background. After 24, 48, 72, 96, and 120 h of inoculation, the medium in the wells to be determined was discarded; after the medium containing 100 µL of 10% CCK-8 (Dojindo Laboratories) was added, the mixture was cultured in an incubator at 37 °C and 5% CO2 for 2 h. Absorbance was measured spectrophotometrically (BioTek Instruments, Inc) at 450 nm.
Measurement of half maximal inhibitory concentration (IC50)
Optimal number of cells were inoculated in 96-well plates; then, 100 µL of cell suspensions were prepared. After the cells were plated and then became adherent, 100 µL of drug-containing medium at different concentration gradients (gemcitabine: 0–1,000 nM) were added to the wells. The medium containing DMSO was used as the 0 nM blank. After the culture plate was inoculated in the incubator for 24 h, the culture medium was pipetted away and 100 µL of medium containing 10% CCK-8 was added. The culture plate was incubated in the incubator for 1–4 h before the optical density (OD) of the cells was measured. With a detection wavelength of 450 nm, the OD of the cells was measured on a micro-plate reader. The survival rate of the cells was calculated based on the relative ratio of average OD values in each group. Based on the cell survival rate and the matched drug concentration, curve fitting with nonlinear regression was performed by using the GraphPad Prism 6.0 (San Diego, CA, USA) to obtain the IC50.
Western blotting
Western blotting was performed to determine the expression levels of different proteins in TNBC cells. Cells were harvested, washed with cold 1×PBS, lysed with RIPA lysis buffer (Beyotime, Shanghai, China) for 30 min on ice, and then centrifuged at 15,000 g for 10 min at 4 °C. The total protein concentration was determined using a Bio-Rad protein Assay Kit (Hercules, CA, USA). Equal amounts (20 µg per load) of protein samples were subjected to SDS-PAGE electrophoresis and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Darmstadt, Germany). The blots were blocked in 10% non-fat milk and incubated with primary antibodies, followed by incubation with secondary antibodies conjugated with horseradish peroxidase (HRP). Antibodies to β-actin, p21, SOD2 and γH2A.X were purchased from Abcam (Cambridge, MA, USA). The secondary antibodies were the F(ab)2 fragment of donkey anti-mouse immunoglobulin (product NA931) or the donkey anti-rabbit immunoglobulin (product NA9340) linked to horseradish peroxidase and were obtained from Amersham Biosciences (Little Chalfont, Buckinghamshire, UK).
Array-based gene expression profiling
The total RNA of samples was extracted and then analyzed on the Aglient 2100 Bioanalyzer (Aglient). The amplified RNA (aRNA) was prepared by using the GeneChip 3’IVT Express Kit (Affymetrix). After the synthesis of the first strand cDNA, synthesis of the second strand was performed for double-stranded DNA; then, biotin-labeled aRNA was obtained by in vitro reverse transcription. After the aRNA was purified, it was fragmented and then underwent chip probe hybridization on the GeneChip Hybridization Oven 645 (Affymetrix). Upon the completion of the hybridization, the arrays were washed and stained with the GeneChip Fluidics Station 450. Finally, the graphs and raw data were scanned and obtained on the GeneChip Scanner 3000 (Affymetrix). For data processing, raw data were normalized using the oligo package in R with Robust Multi-array Analysis (RMA). We performed Bayes statistics using empirical Bayes (eBayes) in the limma package in R to calculate which probes were significantly differentially expressed between MDA-MB-231 and 231-RR (or between MDA-MB-231 and 231-GEM). We used Fisher’s exact test to verify whether a transcript was indeed significantly differentially expressed. Differentially expressed gene were characterized according to the following criteria: an empirical foldchange greater than 1.5 and an eBayes test P value less than 0.05.
Construction and packaging RNAi lentivirus vector
The restriction endonucleases were used to digest and obtain the linearized vector. (I) vector name: GV248; (II) component form: hU6-MCS-Ubiquitin-EGFP-IRES-puromycin; (III) control number: CON077; (IV) insertion sequence of the control: TTCTCCGAACGTGTCACGT. All these plasmids were purchased from GeneChem (Shanghai, China). The target fragments were prepared by primer annealing. The designed primers were added with the enzyme-cutting sites at both of its ends. The paired primer powder was dissolved in the annealing buffer solution, water bathed at 90 °C for 15 min, and then cooled down to room temperature. After the primer was annealed, it contained the same enzyme-cutting sites as the two ends of the linearized cloning vector. A reaction system was prepared with the linearized vector and annealing product for ligation, whose product was directly transformed. Single clone on the plate was selected for PCR identification, and the positive clones were sequenced and analyzed. The proper bacterial solution was submitted for amplification culture and extraction to yield high-purity plasmid for virus packaging. The 293T cells were co-transfected with three plasmids. Virus (i.e., the unpurified cell culture supernatant) was harvested 48–72 h after the completion of transfection. According to the requirement of the experiment, the high-titer lentivirus preservation solution was obtained after concentration and purification. Finally, each indicator of the lentivirus was strictly determined before the virus was used to infect the target cells. After the puromycin-labeled lentivirus infected the cells for 48–72 h, the cells were screened with puromycin for 48 h, and cells with a confluence rate of 70–80% were harvested.
Induction of radioresistant tumor xenograft in nude mice
Accordance to the established research protocol, a certain dose of radiation was used to repeatedly irradiate the tumor xenograft from nude mice with MDA-M-231 to induce radioresistant tumor xenografts, which were preserved for further experiments. This procedure was based on our clinical observations and experiences. First, only radioresistant tumor xenografts that had survived after high-dose irradiation (i.e., tumor tissues that could continue to be transplanted into nude mice and formed tumors) were used in this experiment. Second, in the later period of X-ray induction, the re-tumorigenesis and growth of the tumor xenografts dramatically slowed down after repeated irradiation (compared with the control). The mice experiment protocol was approved by the Animal Ethics Committee of Shanghai Medical College of Fudan University. During the experiments, all the animals were handled according to the AAALAC requirements.
Immunohistochemical staining
Tissues were dehydrated in graded ethanol solutions, cleared in 3 changes of xylene, and penetrated in heated paraffin. The tissues were embedded in paraffin, cut into 4 to 6 mm sections, and placed onto slides. Before staining, deparaffinization and rehydration were performed. Antigen retrieval was performed using a pressure cooker. The slides were incubated in 1× target retrieval solution (Beyotime, Shanghai, China) at 120 °C for 4 min at 18 to 20 psi. Endogenous hydrogen peroxidase activity was blocked with hydrogen peroxide for 10 minutes, followed by rinsing with wash buffer (Beyotime, Shanghai, China). The slides were incubated with the appropriate antibodies. The antibodies to p21 and SOD2 were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). The secondary antibodies against mouse or rabbit IgG were supplied in an IHC kit from Abcam (Cambridge, MA, USA).
Breast cancer gene expression profile in TCGA database and clinical data
The UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu/) offers relatively complete and reliable from The Cancer Genome Atlas (TCGA) (17). The breast cancer gene expression profile data used in this study were downloaded from this website. Access date: 2015-02-24: Label: TCGA breast invasive carcinoma (BRCA) gene expression by RNAseq (IlluminaHiSeq); Authors: University of North Carolina TCGA genome characterization center; Dataset ID: TCGA_BRCA_exp_HiSeqV2. Totally 1,215 cases of invasive breast cancer were identified, and the downloaded gene expression data and clinical data were further processed according to the requirements of statistical analysis. The gene expression information and the clinical data downloaded from the database were strictly matched according to the sequence number of the TCGA sample. The results were verified by two independent reviewers to ensure the accuracy of data.
Data processing and statistical analysis
Statistical analyses were performed by using the IBM SPSS 22.0 (IBM SPSS, NY, USA). The continuous data were expressed as mean ± standard deviation. Different groups’ comparisons were performed by using a one-way ANOVA, whereas paired comparisons were based on LSD t-test; for continuous data containing repeated measurements, two-way ANOVA were performed. The cut-off values of gene expressions in breast cancer patients in TCGA database were determined by using X-tile software (18); alternately, the medians of gene relative expression levels were used to stratify the expression level of a specific gene in the dataset into low expression and high expression. Of the clinical data of breast cancer patients in the TCGA database, the survival was analyzed by using the Kaplan-Meier method, and the comparisons of two survival curves were based on log-rank test. Univariate Cox regression was used for the initial screening of prognostic factors, and then variables with a P<0.10 in the univariate analysis were enrolled in the multivariate Cox regression model. During the multivariate regression analysis, P<0.05 was considered statistically significant.
Results
Induction and verification of radioresistant TNBC lines
We used the CCK-8 kit to detect the proliferation ability of parental 231 cells and radioresistant 231-RR cells; as shown in Figure 1C, the proliferation rate of 231-RR cells was significantly lower than that of 231 cells. Compared with the parental 231 cells, the radioresistant 231-RR cells show radioresistant properties: the SF2 =0.574 in 231 and SF2 =0.740 in 231-RR (Figure 1D). Figure 1E shows the survival curve of the 231-RR cells after five passages and that of the parental 231 cells: the SF2 was 0.601 in 231 cells and 0.675 in 231-RR, showing remarkably weakened radioresistance in the resistant strain. Figure 1F shows the cell survival curve of 231-RR cells and parental 231 cells after maintenance irradiation at low dose (2 Gy per fraction once weekly): the SF2 was 0.579 in 231 cells and 0.753 in 231-RR. Thus, maintenance irradiation at low dose (2 Gy/Fx/week) is an effective way for maintaining the stable radioresistance of cell line. Therefore, we successfully constructed the radioresistant 231-RR cells, whose radioresistance could be maintained at low dose (2 Gy/Fx/week).
Verification of the drug resistance of TNBC gemcitabine-resistant lines
We verified the resistance of TNBC gemcitabine-resistant strain (231-GEM), and its parental 231 cells by determining their IC50. As shown in Figure 2A,B, the IC50 was 2.19 nM in the parental 231 cells, which was significantly lower than that (IC50 =19.79 nM) in 231-GEM cells. We further verified the proliferation ability of the parental 231 cells and gemcitabine-resistance of 231-GEM cells by using the CCK-8 kit. As shown in Figure 2C, the proliferation ability of gemcitabine-resistant strain had significantly decreased when compared with the parental 231 strain. We determined the clonogenic abilities of parental strain and gemcitabine-resistant strain by performing the clonogenic assay. After 2 weeks of co-culture of 2 µM of gemcitabine with cells, the count of clones was compared between 231-GEM strain and parental 231 cells. As shown in Figure 2D,E,F, the count of clones was significantly higher in 231-GEM strain than in parental 213 cell.

Array-based gene expression profiling for MDA-MB-231, MDA-MB-231-RR and MDA-MB-231-GEM
Array-based gene expression profiling was performed for three pairs of 231 cells and 231-RR cells. Differential expressed genes with an absolute fold change greater than 1.5 and a P value <0.05 were regarded as significantly different. A total of 760 differential genes were obtained, of which 356 were up-regulated and 404 were down-regulated. The volcano plot, scatter plot, and cluster diagram of the differential genes in two cell lines are shown in Figure 3A,B,C. We further performed array-based gene expression profiling on three gemcitabine-resistant 231-GEM cell samples, and compared the results with those of three previously detected parental 231 cell samples. A total of 1,321 differential genes were obtained, of which 661 were up-regulated and 660 were down-regulated. The volcano plot, scatter plot, and cluster diagram of the differential genes in two cell lines are shown in Figure 3D,E,F.
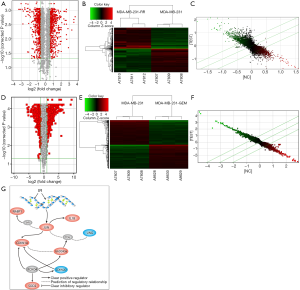
Screening of differential genes in 231-RR cells, 231-GEM cells, and parental MDA-MB-231 cells
We analyzed the comparison result between gemcitabine-resistant 231-GEM and 231 cells and that between 231-RR and 231 cells, and the datasets of differential genes were extracted. The differential gene datasets contained 317 genes, and more detailed information including gene names and fold change is shown in http://cdn.amegroups.cn/static/application/09bb7520d009e0b7409313a95259369a/10.21037atm.2019.12.114-1.pdf. Based on the differential genes overlap screened, and the conclusions of previous researches (19-22), we finally identified the differential expressed genes (Figure 3G), which are mainly involved in DNA repair and cell cycle.
Functional verification of differential expressed genes at cellular level
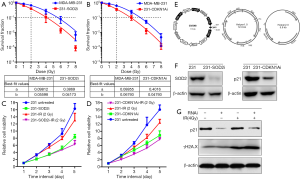
In Figure 4A,B,C,D, after the SOD2 gene was silenced in the 231-cell line, the proliferation of the 231-cell line was remarkably slowed down, along with significantly decreased clonogenic ability. The SF2 was 0.657 in the parental 231 cell line and 0.391 in 231-SODi cell line (Figure 4A,C). After the CDKN1A gene was silenced in the 231-cell line, the proliferation of the 231-cell line was also remarkably slowed down, along with significantly decreased clonogenic ability. The SF2 was 0.638 in the parental 231 cell line and 0.370 in 231-CDKN1Ai cell line (Figure 4B,D). We constructed RNAi lentivirus vectors for SOD2 and CDKN1A (Figure 4E), respectively. Figure 4F shows the expression levels of SOD2 and p21 proteins in the gene knockdown cell lines 231-SOD2i, 231-CDKN1Ai. Obviously, the viral vectors had a good effect on the SOD2 and CDKN1A genes. Figure 4G shows the protein expressions in parental 231 cells and 231-CDKN1Ai cells after 2 h of 4-Gy X-ray irradiation, in which p21 (CDKN1A gene encoding protein) is a negative cell-cycle regulation protein; and the silencing of CDKN1A gene enhanced the DNA damage after irradiation, and the expression of γH2A.X increased accordingly.
Induction of radioresistant tumor xenografts in MDA-MB-231
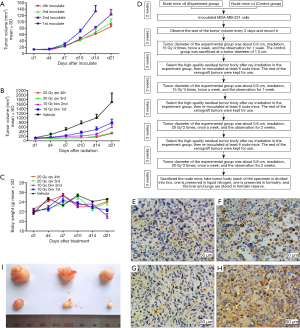
We transplanted the high-quality tumor xenografts that were selected after the first three IR induction cycles. The tumorigenesis of these xenografts was significantly slower than in the previous three cycles (Figure 5A,B,C,D). For non-irradiated tumor xenografts, tumorigenesis occurred typically 2 weeks after transplantation, with a tumor size of 133.4±18.4 mm3; for irradiated tumor xenografts, the tumorigenesis slowed down and typically needed 3 weeks to reach the required size; the tumor sizes ranged 85.4±7.5 mm3 before the fourth cycle (Figure 5A). During the 4th induction cycle, the growth rate of the irradiated tumor xenografts was also lower than those in the previous induction cycles (Figure 5B). In all the radiation groups, the body weight of the nude mice showed no significant change throughout the experiment (Figure 5C).
Expression levels of differentially expressed genes in radioresistant tumor xenografts
Three tumor xenografts were randomly selected from each group for IHC to determine the expression levels of SOD2 and p21 proteins (Figure 5E,F,G,H,I). Figure 5E,F show the SOD2 expression in the control group and radioresistant tumor xenografts, respectively. It is obvious that the SOD2 expression level significantly up-regulated in the radioresistant tumor xenografts. Figure 5G,H show the expression levels of p21 in the control and the radioresistant tumor xenografts, and the p21 expression was significantly up-regulated in the radioresistant tumor xenografts. IHC performed in nude mice further verified the results of cellular results: the up-regulated SOD2 and CDKN1A were associated with the radioresistance of TNBC cells.
Verification of differential gene expression in TCGA database
The cut-off value for SOD2 was 11.39 (the relative expression level of the gene after standardization), with 9.01–11.39 being the SOD2 low-expression group and 11.39–16.02 being the SOD2 high-expression group. The cut-off value for CDKN1A was 11.17, with 6.79–11.17 being the CDKN1A low-expression group and 11.17–14.48 being the CDKN1A high-expression group. The baseline clinical features of 1,215 patients with breast cancer are shown in Table 1.
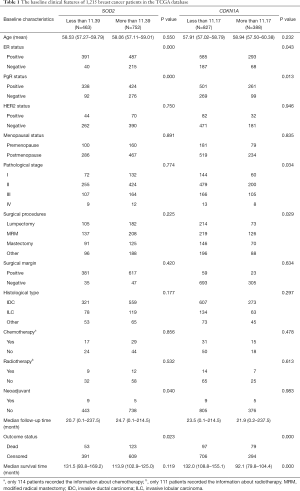
Full table
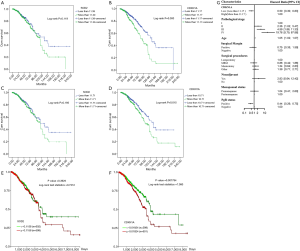
The survival analysis 1,215 patients with breast cancer is shown in Figure 6. Figure 6A shows the survival curves of breast cancer patients with low SOD2 expression and high SOD2 expression. The median survival time was 131.5 months (range: 93.8–169.2 months) in patients with low SOD2 expression and 113.93 months (range: 102.9–125.0 months) in those with high SOD2 expression; thus, the prognosis of patients with low SOD2 expression was superior to those with high SOD2 expression, although the difference was not statistically significant (log-rank P=0.119). Figure 6B shows the survival curves of patients with high or low CDKN1A expression. The median survival time was 132.0 months (range: 108.8–155.1 months) in patients with low CDKN1A expression and 91.1 months (range: 79.8–104.4 months) in those with high CDKN1A expression. Thus, the prognosis was significantly better in patients with low CDKN1A expression than in those with high CDKN1A expression (log-rank P=0.000). The above conclusion was based on the statistical results obtained by determining the optimal cutoffs in the X-tile software. When we further divided the patients into high and low SOD2/CDKN1A expression groups based on the medians of the relative expressions of the SOD2 and CDKN1A genes, we obtained the similar results (Figure 6C,D). By using a web-based survival analysis tool (http://xena.ucsc.edu/), we divided the medians of the SOD2 and CDKN1A expression levels into two groups for log-rank test, which showed similar results (Figure 6E,F).
The Cox regression analysis showed that the expression level of CDKN1A gene was an independent prognostic factor for breast cancer patients (P=0.008). The calculation results are shown in Table 2 and Figure 6G, among which Table 2 summarizes the results of univariate analysis and multivariate Cox regression analysis, and Figure 6G is the forest plot of multivariate regression analysis.
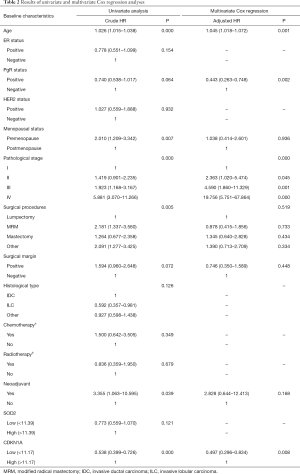
Full table
Discussion
We constructed the radioresistant MDA-MB-231-RR cell line, and found that continuous low doses of X-ray irradiation (2 Gy/Fx/week) can better maintain the stability of radioresistance. Totally 163 up-regulated genes and 156 down-regulated genes were obtained. Two genes (SOD2 and CDKN1A) that were up-regulated in radioresistant and gemcitabine-resistant cell lines were selected from the overlap of differential expressed genes and then verified their functions in cells and nude mice tumor xenografts. Cellular results showed that the radiosensitivity of the MDA-MB-231 cell line was significantly enhanced after the knockdown of SOD2 and CDKN1A genes. Meanwhile, we tried to apply a novel radioresistant tumor xenograft induction mode in nude mice and obtained radioresistant breast cancer tumor xenograft, which was used for verifying the differential expressed genes obtained during the previous screening. The expression of SOD2 and CDKN1A encoding proteins increased in radioresistant MDA-MB-231 xenografts, demonstrating SOD2 and CDKN1A genes mediate the radioresistance of TNBC cell line. Based on the expression levels of SOD2 and CDKN1A genes, we divided 1,215 breast cancer patients in TCGA database into high and low SOD2/CDKN1A expression groups. The prognosis of the low SOD2 expression group was superior to that of the high SOD2 expression group, although the difference was not statistically significant. In contrast, the prognosis of the low CDKN1A expression group was significantly superior to that of the high CDKN1A expression group. We found that the expression level of CDKN1A gene was an independent prognostic factor in breast cancer patients.
It is generally believed that SOD2 is associated with the repair of mitochondrial damage. As shown in our current research, the up-regulated expression of SOD2 gene in radioresistant or drug-resistant TNBC cell line demonstrates SOD2 is involved in the mediation of radioresistance or drug-resistance, and cell lines with high SOD2 expression may have a better ability to resist stress and damage. Verification of the data from TCGA database demonstrated that breast cancer patients with high SOD2 expression had worse prognosis than those with low SOD2 expression, although the difference was not statistically significant. Chang et al. found that, in patients with adenoid cystic carcinoma of the parotid gland, high SOD2 expression was associated with higher distant metastasis and lower overall survival and disease-free survival; they concluded that salivary adenoid cystic carcinoma with high SOD2 expression has higher invasion and metastasis abilities and thus has a worse prognosis (23). In 258 cases of esophageal squamous cell carcinoma, Ma et al found that patients with high HOXA13, ANXA2, and SOD2 gene expressions had shorter survival time and worse prognosis (24). In addition, the role of the SOD2 gene in cancer progression is also multifaceted. In Wang et al.’s study, the overexpression of Hexokinase 2 enhanced the invasion and metastasis of tongue cancer by activating the SOD2-H2O2 pathway and thus increasing the SOD2 activity (25). Zhang et al. investigated the overexpressed SOD2 gene in colon cancer cell by using gene technology. It was found that overexpression of SOD2 could significantly increase the radiosensitivity of colon cancer cells and protect normal cells (26). Despite these contradictions, we conclude, based on our current findings and previous literature, that SOD2 is a “double-edged sword”. While high SOD2 expression can protect normal tissues, it can also make the malignant tumor tissue become resistant to radiotherapy or chemotherapy, leading to treatment failure.
The up-regulated expression of CDKN1A gene in radioresistant and drug-resistant TNBC cell lines suggests that these cell lines have enhanced ability to repair the damage caused by X-ray or chemotherapeutic drugs. Verification with the data in the TCGA database also showed that the prognosis of breast cancer patients with high CDKN1A expression had significantly worse prognosis than that of patients with low CDKN1A expression. However, the findings varied in previous literatures. Nevertheless, the conclusions were more consistent among reports on the relationship between p21 and tumor cell radiosensitivity. Huerta et al. have found that the p21 and Bax gene deletion is associated with the radiosensitization of colon cancer (27). Hötte et al. discovered the overloading of RNPC1 gene in the esophageal adenocarcinoma cell line could up-regulate p21 expression and thus further mediate the intracellular radioresistance (28). Yang et al. concluded that RNA interference-mediated knockdown of p21 could inhibit the cell cycle arrest at G2/M phase caused by targeted drug and thus enhance the radiosensitivity (29). Graham et al found that FAK could inhibit p21 and thus increase the radiosensitivity of squamous cell carcinoma cells in rat models (30).
Our current research had the following innovations: First, we constructed a radioresistant TNBC cell model and explored the method for maintaining stable radioresistance property. Previous studies have described a variety of methods for inducing and constructing radioresistant cell lines (12,13). However, while these methods are practically feasible in inducing radioresistance, how to maintain the stable inheritance of such radioresistance remains challenging. In our current study, we maintained the stable resistance in the radioresistant cell lines by maintaining low-dose irradiation at a dose of 2 Gy/Fx/week, which may also shed light on future studies on maintaining radioresistance in other cell lines. Second, we explored a new mode for inducing radioresistant tumor xenografts, which had not been reported in the literature before. Such a mode may also be used in other radiobiological studies on tumors. Third, we found that SOD2 and CDKN1A genes could be used as markers for predicting the radiosensitivity of TNBC. Fourth, we further verified the experimental results at cell and animal levels with the data of 1,215 breast cancer patients in the TCGA database and found that the expression level of CDKN1A gene was an independent prognostic factor in breast cancer patients.
Despite the rigorous experimental design based on the existing conditions, our study still had the following limitations: First, no definite criteria have been available for determining the presence of radioresistance in repeatedly irradiated tumor xenografts during the induction of radioresistant tumor xenografts. Second, there are some inherent limitations when the breast cancer samples from the public TCGA database were used for verification: (I) these patients were followed up for a short period of time. The median follow-up duration was only 25.45 months. Therefore, many patients did not develop the endpoint events we defined. These censored data limited the statistical power of our study; (II) we did not strictly distinguish and further screen TNBC samples from non-TNBC samples in the TCGA database; instead, we selected all the invasive breast cancer samples for verification. This was because many samples in the TCGA database lack data on the ER, PgR, and HER2 status and therefore it is difficult to carry out re-evaluation. Similarly, the information on chemotherapy, radiotherapy, and/or endocrinology was incomplete in these 1,215 patients, and most treatment-related data were absent; (III) when we analyzed the prognosis of 1,215 breast cancer patients in the TCGA database, Cox regression analysis showed that HER2 was not an independent prognostic factor in these patients. However, HER2 is generally believed to be an important factor affecting the prognosis of breast cancer patients.
Conclusions
It is therefore concluded that: (I) it is feasible to conduct radiobiologic research by constructing radioresistant cell lines and inducing radioresistant tumor xenografts. (II) SOD2 and CDKN1A genes mediate the radioresistance of TNBC. (III) High expressions of SOD2 and CDKN1A are associated with poor prognosis in breast cancer. These two genes can be used as predicted markers to determine the radiosensitivity of breast cancer.
Acknowledgments
Funding: This work was partly financially supported by the National Natural Science Foundation of China (Grant Numbers: 81072164, 81372430, 81402525, and 81602668), Shanghai Municipal Commission of Health and Family Planning Grant (Grant Number: 201440425), the Municipal Natural Science Foundation of Shanghai (Grant Number: 16ZR1406900) and the Shanghai Youth Medical Talents-Specialist Program.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Clarke M, Collins R, Darby S, et al. Effects of radiotherapy and of differences in the extent of surgery for early breast cancer on local recurrence and 15-year survival: an overview of the randomised trials. Lancet 2005;366:2087-106. [Crossref] [PubMed]
- McGale P, Taylor C, Correa C, et al. Effect of radiotherapy after mastectomy and axillary surgery on 10-year recurrence and 20-year breast cancer mortality: meta-analysis of individual patient data for 8135 women in 22 randomised trials. Lancet 2014;383:2127-35. [Crossref] [PubMed]
- Bauer KR, Brown M, Cress RD, et al. Descriptive analysis of estrogen receptor (ER)negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype - A population-based study from the California Cancer Registry. Cancer 2007;109:1721-8. [Crossref] [PubMed]
- Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin Cancer Res 2007;13:4429-34. [Crossref] [PubMed]
- Cleator S, Heller W, Coombes RC. Triple-negative breast cancer: therapeutic options. Lancet Oncol 2007;8:235-44. [Crossref] [PubMed]
- Chen X, Yu X, Chen J, et al. Analysis in early stage triple-negative breast cancer treated with mastectomy without adjuvant radiotherapy: patterns of failure and prognostic factors. Cancer 2013;119:2366-74. [Crossref] [PubMed]
- Chen X, Yu X, Chen J, et al. Radiotherapy can improve the disease-free survival rate in triple-negative breast cancer patients with T1-T2 disease and one to three positive lymph nodes after mastectomy. Oncologist 2013;18:141-7. [Crossref] [PubMed]
- Brosnan EM, Anders CK. Understanding patterns of brain metastasis in breast cancer and designing rational therapeutic strategies. Ann Transl Med 2018;6:163. [Crossref] [PubMed]
- Kyndi M, Sorensen FB, Knudsen H, et al. Estrogen receptor, progesterone receptor, HER-2, and response to postmastectomy radiotherapy in high-risk breast cancer: the Danish Breast Cancer Cooperative Group. J Clin Oncol 2008;26:1419-26. [Crossref] [PubMed]
- Hou J, Zhou Z, Chen X, et al. HER2 reduces breast cancer radiosensitivity by activating focal adhesion kinase in vitro and in vivo. Oncotarget 2016;7:45186-98. [Crossref] [PubMed]
- Pearce AG, Segura TM, Rintala AC, et al. The generation and characterization of a radiation-resistant model system to study radioresistance in human breast cancer cells. Radiat Res 2001;156:739-50. [Crossref] [PubMed]
- Tyrsina EG, Slanina SV, Kakpakova ES, et al. Isolation and characterization of highly radioresistant malignant hamster fibroblasts that survive acute gamma irradiation with 20 Gy. Radiat Res 2005;164:745-54. [Crossref] [PubMed]
- Ye FG, Song CG, Cao ZG, et al. Cytidine Deaminase Axis Modulated by miR-484 Differentially Regulates Cell Proliferation and Chemoresistance in Breast Cancer. Cancer Res 2015;75:1504-15. [Crossref] [PubMed]
- Scott JG, Berglund A, Schell MJ, et al. A genome-based model for adjusting radiotherapy dose (GARD): a retrospective, cohort-based study. Lancet Oncol 2017;18:202-11. [Crossref] [PubMed]
- Torres-Roca JF, Fulp WJ, Caudell JJ, et al. Integration of a Radiosensitivity Molecular Signature Into the Assessment of Local Recurrence Risk in Breast Cancer. Int J Radiat Oncol Biol Phys 2015;93:631-8. [Crossref] [PubMed]
- Zhang G, Wang Y, Chen B, et al. Characterization of frequently mutated cancer genes in Chinese breast tumors: a comparison of Chinese and TCGA cohorts. Ann Transl Med 2019;7:179. [Crossref] [PubMed]
- Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004;10:7252-9. [Crossref] [PubMed]
- Hildesheim J, Bulavin DV, Anver MR, et al. Gadd45a protects against UV irradiation-induced skin tumors, and promotes apoptosis and stress signaling via MAPK and p53. Cancer Res 2002;62:7305-15. [PubMed]
- Qiu X, Brown K, Hirschey MD, et al. Calorie Restriction Reduces Oxidative Stress by SIRT3-Mediated SOD2 Activation. Cell Metabolism 2010;12:662-7. [Crossref] [PubMed]
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009;9:400-14. [Crossref] [PubMed]
- Calaf GM, Hei TK. Ionizing radiation induces alterations in cellular proliferation and c-myc, c-jun and c-fos protein expression in breast epithelial cells. Int J Oncol 2004;25:1859-66. [PubMed]
- Chang B, Yang H, Jiao Y, et al. SOD2 deregulation enhances migration, invasion and has poor prognosis in salivary adenoid cystic carcinoma. Sci Rep 2016;6:25918. [Crossref] [PubMed]
- Ma RL, Shen LY, Chen KN. Coexpression of ANXA2, SOD2 and HOXA13 predicts poor prognosis of esophageal squamous cell carcinoma. Oncol Rep 2014;31:2157-64. [Crossref] [PubMed]
- Wang W, Liu Z, Zhao L, et al. Hexokinase 2 enhances the metastatic potential of tongue squamous cell carcinoma via the SOD2-H2O2 pathway. Oncotarget 2017;8:3344-54. [Crossref] [PubMed]
- Zhang Z, Lang J, Cao Z, et al. Radiation-induced SOD2 overexpression sensitizes colorectal cancer to radiation while protecting normal tissue. Oncotarget 2017;8:7791-800. [PubMed]
- Huerta S, Gao X, Dineen S, et al. Role of p53, Bax, p21, and DNA-PKcs in radiation sensitivity of HCT-116 cells and xenografts. Surgery 2013;154:143-51. [Crossref] [PubMed]
- Hötte GJ, Linam-Lennon N, Reynolds JV, et al. Radiation sensitivity of esophageal adenocarcinoma: the contribution of the RNA-binding protein RNPC1 and p21-mediated cell cycle arrest to radioresistance. Radiat Res 2012;177:272-9. [Crossref] [PubMed]
- Yang D, Tan M, Wang G, et al. The p21-dependent radiosensitization of human breast cancer cells by MLN4924, an investigational inhibitor of NEDD8 activating enzyme. PLoS One 2012;7:e34079. [Crossref] [PubMed]
- Graham K, Moran-Jones K, Sansom OJ, et al. FAK deletion promotes p53-mediated induction of p21, DNA-damage responses and radio-resistance in advanced squamous cancer cells. PLoS One 2011;6:e27806. [Crossref] [PubMed]


