
Complicated paroxysmal kinesigenic dyskinesia associated with SACS mutations
Introduction
Autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS, OMIM #270550) is an early-onset progressive disorder that mainly presents with spinocerebellar ataxia, upper motor neuron dysfunction, and distal sensorimotor and peripheral neuropathy (1). ARSACS is caused by pathogenic variants in SACS (2). Paroxysmal kinesigenic dyskinesia (PKD, OMIM #128200) is characterized by recurrent and brief attacks of dystonia and choreoathetosis lasting no more than 1 minute, triggered by sudden movements (3). Mutations in PRRT2 and SCN8A are associated with PKD (4,5). We describe here two independent cases bearing SACS mutations presented with ARSACS and PKD, which expands the clinical phenotype associated with SACS mutations to include PKD.
Methods
The study was approved by the ethical review board of Peking Union Medical College Hospital. Both families provided written informed consent for clinical-genetic correlation studies.
Genetic analysis
Genomic DNA was extracted from peripheral blood leukocytes using a QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) according to standard protocols. Mutations in PRRT2 were excluded by Sanger sequencing, and WES was then performed on the two patients. Exonic regions were captured and enriched using an Agilent SureSelect Human All Exon 50 Mb kit (Agilent, Santa Clara, CA, USA). The captured fragments were purified and sequenced on a Hiseq2000 platform (Illumina, San Diego, CA, USA) using 90-bp paired-end reads. The sequence was aligned to the human reference genome (UCSC hg19) using a Burrows-Wheeler Aligner (6). The aligned sequence files were reformatted using SAMtools (7). Subsequent annotation was performed using SeattleSeq Annotation 138 (http://snp.gs.washington.edu/SeattleSeqAnnotation138/), SIFT (http://sift.jcvi.org/), and PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/). Variant frequencies were initially determined in dbSNP, the 1000 Genomes Project, and the NHLBI Exome Sequencing Project version ESP6500 exome variant server [http://evs.gs.washington.edu/EVS/ (1 Dec 2013)] to remove common single nucleotide polymorphisms (SNPs). Only nonsynonymous, splicing and frameshift variants with minor allele frequency (MAF) <0.5% or that were absent in population databases were selected for further assessment. De novo mutations suggestive of a dominant disease model or homozygous and compound heterozygous variants of a recessive model were identified and subjected to Sanger sequencing. Genes that may potentially be related to paroxysmal dystonic symptoms were carefully excluded. The WES results of 163 PKD patients without additional symptoms (patients from a previous experiment (8) were also reviewed.
Results
Case reports
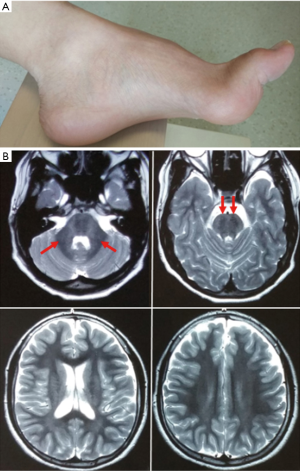
Patient 1 is a 14-year-old male. He developed unsteady gait and poor coordination at approximately 2 years old. Weakness and numbness were also reported, and all the symptoms progressively worsened. At the age of eight, he experienced recurrent and brief attacks of dystonia in unilateral or bilateral limbs triggered by sudden movements, stress or anxiety. The attacks lasted less than 10 seconds and were completely relieved by carbamazepine (CBZ) (200 mg/day). He reported no seizure attacks. Upon physical examination, he had jerky ocular pursuits, saccade dysmetria, mild distal weakness, pyramidal signs, cerebellar ataxia and high arch (Figure 1A).
Patient 2 is a 12-year-old female. She had strangling steps from 1.5 years on that worsened as ataxia became prominent. Mild learning difficulties were also noticed after she had entered primary school. When she was 10 years old, she began to experience episodes of unilateral or bilateral dystonia, causing instant falls with consciousness reserved. These attacks may be triggered by sudden movements or stress. Each attack lasted approximately 5 seconds and was favorably controlled by low-dose CBZ (100 mg/day). Her parents observed no seizure events. Physical examination showed dysarthria, counting deficits, saccade dysmetria, distal weakness, pyramidal signs, and cerebellar ataxia.
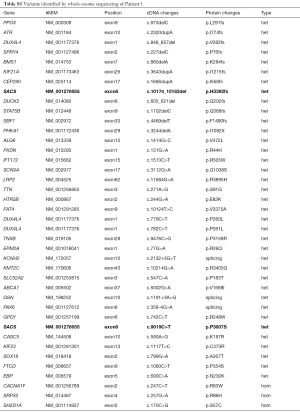
For both patients, routine serum tests, blood lactate concentration, ceruloplasmin, organic acids and amino acids were performed and were all within normal ranges. Neurophysiological examination revealed large-fiber sensorimotor axonal-demyelinating neuropathy. Brain magnetic resonance imaging revealed generalized atrophy, most markedly affecting the cerebellum. Typical cerebellar and pontine changes manifesting as hypointensities in T2-weighted images associated with ARSACS (9) were observed in our patients (see Figure 1B). For both patients, electroencephalography (EEG) detected no epileptic discharges. Increased amounts of slow waves in the bilateral frontal lobe were detected in Patient 2. Neurophysiological examination showed sensorimotor axonal polyneuropathy for both patients.
Genetic tests for spinocerebellar ataxias 1, 2, 3, 6, 7, 8, 10, 12 and 17, Friedreich ataxia, and dentatorubral-pallidoluysian atrophy (DRPLA) were negative, and patients’ DNA samples were subsequently subjected to whole-exome sequencing (WES). After the SACS mutations were identified, both patients were treated with energy supplements primarily to treat mitochondrial deficits. During the 4-year follow up, Patient 1 deteriorated slowly in his motor skills, with mild distant limb numbness and weakness. However, other than that, his ataxia was relatively stable, and his PKD was controlled satisfactorily. Patient 2 was lost to follow-up.
None of the rare variants with a minor allele frequency less than 0.5% were identified in the known genes associated with PKD. As other paroxysmal dystonic disorders cannot be completely excluded, genes potentially associated with the current symptoms were also carefully examined. These included the genes of ADCY5, ATP1A3, GCH1, KCNMA1, PARK2, MR1, SLC2A1, KCNA1, CACNB4, CACNA1A, CACNB4, SLC1A3, KCNQ2 and SLC1A3. One variant was found in CACNA1A (c.1337T>C) in Patient 2. It was inherited from her mother who had no history of episodic symptoms and showed no signs of ataxia upon physical examination. The CACNA1A variant was thus considered less likely to be the causal gene. All potential pathogenic variants of the two patients are listed in Tables S1 and S2.
Full table
Full table
The SACS gene (NM_014363.5) was the only gene associated with the existing phenotype in concordance with the symptoms of Patient 1 and Patient 2 and shared by them. Novel compound heterozygous or homozygous variants were revealed when comparing candidate genes and were confirmed by Sanger sequencing in both patients. Patient 1 holds one missense and one frameshift variant in the SACS gene (c.9019C>T, p.P3007S and c.10174_10183del, p.H3392fs) (Figure 2A). The homozygous variant presented in Patient 2 (c.4127G>A, p.W1376X) (Figure 2B) should be deleterious and would possibly cause truncation of the SACS protein. In both patients, each mutant allele was inherited from one of his or her unaffected parents. All 3 mutations had not been reported before and were absent in 196 ethnic-matched control chromosomes or in data from the 1000 Genomes Project. They were thus assigned as likely pathogenic.
In searching for additional genes that may have caused the PKD symptoms of our patients, we also reviewed the WES results of patients with pure PKD symptoms for SACS variations. There were 34 patients who carried heterozygous variants out of 163 PKD patients. However, 17 of these variants were documented in public databases with a reported allele frequency of over 0.3%. Three patients carried a pathological PRRT2 mutation simultaneously. No homozygous or compound heterozygous variants in the SACS gene were detected in these patients.
Discussion
ARSACS is a neurodegenerative disease resulting from mutations in SACS. Over 170 mutations with diverse phenotypes have been reported worldwide and are thought to cause loss of function of sacsin (1). The typical ARSACS phenotype consists of a childhood-onset triad of cerebellar ataxia, peripheral neuropathy, and pyramidal tract signs. Reports describe patients with atypical features in addition to ataxia and peripheral neuropathy, which include delayed onset ataxia, non-ataxic spastic paraplegia, mild pyramidal signs, cognitive decline, and widespread supratentorial abnormalities (10-13). Seizures including progressive myoclonus epilepsies were also reported (11,14-16). PKD, however, has never been documented in ARSACS before. Commonly, PKD is simple and pure in phenotype in addition to its association with epilepsy and migraines. Herein, we described 2 unrelated patients bearing mutations in SACS who presented with ARSACS and PKD. These findings may indicate that SACS is highly likely the causal gene of the combined syndrome and should be put to future biological validation.
The SACS gene encodes sacsin, a protein located on the mitochondrial surface. Sacs knockout (Sacs-/-) mice displayed an abnormal gait with progressive motor dysfunction. Clinical features were accompanied by an early onset, progressive loss of cerebellar Purkinje cells followed by spinal motor neuron loss and peripheral nerve dysfunctions highly reminiscent of ARSACS (17). Remarkable bioenergetic damage in ARSACS cells is indicated by reduced basal, adenosine triphosphate (ATP)-linked and maximal mitochondrial respiration rates and by reduced respiratory chain activities and mitochondrial ATP synthesis (18). These findings show that defects in mitochondrial dynamics are the underlying pathophysiological basis of ARSACS (19). Supplementation of mitochondrial energy may thus be the current plan for symptom rescue, and it can be expected that in the near future, gene therapy may possibly be the cure.
In conclusion, we identified mutations in the SACS gene in two independent patients with ARSACS and PKD. Patients with SACS mutations may manifest a PKD phenotype under the background of ARSACS. This is different from PKD patients with mutations in PRRT2 or other mutant genes, such as SCN8A, who show relatively pure paroxysmal dystonic deficits.
Acknowledgments
We thank the patients and their family members for collaboration in this report.
Funding: This study was supported by the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (CIFMS) (Grant number: 2016-I2M-1-002; 2016-I2M-1-004), National Natural Science Foundation of China (No. 81571086; 81870889), Shanghai Municipal Education Commision-Gaofeng Clinical Medicine Grant (No. 20161401), and Research Fund from CAAE (UCB-2018029).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the ethical review board of Peking Union Medical College Hospital (No. JS-1049 of PUMCH). Both families provided written informed consent for clinical-genetic correlation studies.
References
- Bouhlal Y, Amouri R, El Euch-Fayeche G, et al. Autosomal recessive spastic ataxia of Charlevoix-Saguenay: an overview. Parkinsonism Relat Disord 2011;17:418-22. [Crossref] [PubMed]
- Engert JC, Berube P, Mercier J, et al. ARSACS, a spastic ataxia common in northeastern Quebec, is caused by mutations in a new gene encoding an 11.5-kb ORF. Nat Genet 2000;24:120-5. [Crossref] [PubMed]
- Ebrahimi-Fakhari D, Moufawad El Achkar C, Klein C. PRRT2-Associated Paroxysmal Movement Disorders. In: Adam MP, Ardinger HH, Pagon RA, et al. editors. GeneReviews®. Seattle (WA), 1993.
- Gardella E, Becker F, Moller RS, et al. Benign infantile seizures and paroxysmal dyskinesia caused by an SCN8A mutation. Ann Neurol 2016;79:428-36. [Crossref] [PubMed]
- Chen WJ, Lin Y, Xiong ZQ, et al. Exome sequencing identifies truncating mutations in PRRT2 that cause paroxysmal kinesigenic dyskinesia. Nat Genet 2011;43:1252-5. [Crossref] [PubMed]
- Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [Crossref] [PubMed]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078-9. [Crossref] [PubMed]
- Tian WT, Huang XJ, Mao X, et al. Proline-rich transmembrane protein 2-negative paroxysmal kinesigenic dyskinesia: Clinical and genetic analyses of 163 patients. Mov Disord 2018;33:459-67. [Crossref] [PubMed]
- Shimazaki H, Takiyama Y, Honda J, et al. Middle cerebellar peduncles and Pontine T2 hypointensities in ARSACS. J Neuroimaging 2013;23:82-5. [Crossref] [PubMed]
- Ali Z, Klar J, Jameel M, et al. Novel SACS mutations associated with intellectual disability, epilepsy and widespread supratentorial abnormalities. J Neurol Sci 2016;371:105-11. [Crossref] [PubMed]
- Nascimento FA, Canafoglia L, Aljaafari D, et al. Progressive myoclonus epilepsy associated with SACS gene mutations. Neurol Genet 2016;2:e83. [Crossref] [PubMed]
- Gregianin E, Vazza G, Scaramel E, et al. A novel SACS mutation results in non-ataxic spastic paraplegia and peripheral neuropathy. Eur J Neurol 2013;20:1486-91. [PubMed]
- Baets J, Deconinck T, Smets K, et al. Mutations in SACS cause atypical and late-onset forms of ARSACS. Neurology 2010;75:1181-8. [Crossref] [PubMed]
- Pilliod J, Moutton S, Lavie J, et al. New practical definitions for the diagnosis of autosomal recessive spastic ataxia of Charlevoix-Saguenay. Ann Neurol 2015;78:871-86. [Crossref] [PubMed]
- Tzoulis C, Johansson S, Haukanes BI, et al. Novel SACS mutations identified by whole exome sequencing in a Norwegian family with autosomal recessive spastic ataxia of Charlevoix-Saguenay. PLoS One 2013;8:e66145. [Crossref] [PubMed]
- Stevens JC, Murphy SM, Davagnanam I, et al. The ARSACS phenotype can include supranuclear gaze palsy and skin lipofuscin deposits. J Neurol Neurosurg Psychiatry 2013;84:114-6. [Crossref] [PubMed]
- Lariviere R, Gaudet R, Gentil BJ, et al. Sacs knockout mice present pathophysiological defects underlying autosomal recessive spastic ataxia of Charlevoix-Saguenay. Hum Mol Genet 2015;24:727-39. [Crossref] [PubMed]
- Criscuolo C, Procaccini C, Meschini MC, et al. Powerhouse failure and oxidative damage in autosomal recessive spastic ataxia of Charlevoix-Saguenay. J Neurol 2015;262:2755-63. [Crossref] [PubMed]
- Girard M, Lariviere R, Parfitt DA, et al. Mitochondrial dysfunction and Purkinje cell loss in autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS). Proc Natl Acad Sci U S A 2012;109:1661-6. [Crossref] [PubMed]


