
Single-cell RNA profiling of glomerular cells in diabetic kidney: a step forward for understanding diabetic nephropathy
Transcriptomic profiling of kidney tissues or isolated glomeruli has been used to examine changes in the expression of multiple genes in diabetic kidney disease (1,2). Bulk tissue RNA-sequencing can quantitate the average expression of genes in a given tissue but the amount of cell type specific gene expression can only be roughly inferred by the different cell type proportions in each tissue. Accordingly, in these averaged data sets, it is difficult to dissect cell-to-cell variations and identify specific cells that may play an essential role in disease development and progression. The recent and rapid development of single-cell RNA sequencing (scRNA-seq) methods is enabling cell type-specific transcriptome profiling (3-5). This new technique has led to a paradigm shift in the field of genomics, away from bulk tissue analysis, and towards detailed and comprehensive studies of individual cells from different organs and tissues (3). Of importance to the kidney field is a recent report by Fu et al. in the Journal of American Society of Nephrology (6). This paper describes the specific scRNA-seq analyses of isolated glomerular cells from mice with streptozotocin-induced diabetes crossed with endothelial nitric oxide synthase (eNOS)-deficient (eNOS−/−) mice (6). This is a diabetic model with severe endothelial dysfunction and hypertension owing to eNOS deficiency of DKD (7,8). Non-diabetic control eNOS−/− mice were also studied but unfortunately non-diabetic wild type controls were not used (6). The study is nevertheless important for two main reasons: (I) previous scRNA-seq analyses have been focused in the study of gene expression profiles in normal kidneys, without evaluating pathological kidneys, and (II) the majority of renal transcriptomic profiling have been performed using whole kidney tissues, where almost 80% of the cells are tubular cells (9). It would be interesting to perform similar studies in models of DKD other than the one induced by STZ administration.
Gene expression profiling is a widely used approach to examine molecular mechanisms underlying pathological processes. Until now whole tissue and organs consisting of several cell types for gene expression studies have been generally used. Some organs, and particularly the kidneys, however, are endowed with a variety of cell types. The majority of representative cells in bulk studies of kidney tissue moreover reflect largely tubular cells that account for about 90% of the kidney parenchyma. Therefore a transcriptome based on a whole kidney tissue sample is difficult to interpret. The study of single cells transcriptome from tissues has been elusive for years (10), and the first kidney single cells transcriptome was performed from cultured cells (11). During the last decade, microarray technology has been developed for gene expression analysis, and the single-cell cDNA amplification and analyzation has become a reality (5,10,12). With this new technology novel genes and several key signaling pathways in different organs and tissues have been identified (5,10,12).
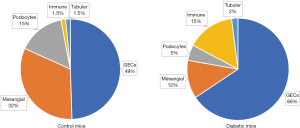
A kidney cell atlas has been extensively used and is constantly updated to examine the cell-specific expression of various proteins in human tissues. This is a valuable tool that complements specific studies in the kidney field from investigators interested in their protein and cell type of interest (13). The studies to identify cell types based on immune staining of proteins unique to a specific cell type are at best semi-quantitative. An important study by Sustzak group provided a molecular definition of cell types in the whole mouse kidney using scRNA-seq of 57,979 cells (9). This study distinguished 21 major cell types by quantitative gene expression that included almost the majority of previous already described cell types, novel transitional cells in the collecting duct and two other undefined cells (9). Focusing on the mouse glomerulus, Karaiskos et al. (13) used glomeruli isolated by dynabeads, for preparation of a single-cell suspension and subsequent scRNA-seq. Their data analysis was able to identify five major cell clusters form 12,956 cells. Based on marker genes, these cluster cells corresponded to glomerular endothelial cells, mesangial cells, podocytes, immune cells, and tubular cells (13). However, this study was limited to normal kidneys. The recent JASN study performed glomerular scRNA-seq in an experimental model of diabetic kidney disease (6). These authors found the same cluster cells that were already described previously, corresponding to five type of cells within the glomeruli, both in diabetic and control mice. Interestingly, the proportion of cells were clearly different in both groups as outlined in Figure 1. A higher number of glomerular endothelial cells were detected in diabetic glomeruli in comparison to the control ones. In addition, lower mesangial and podocyte cells were observed in diabetic glomeruli in comparison to control mice (6). In concordance with these findings, the decrease in the number of podocytes in experimental diabetic nephropathy has been consistently described as a marker of glomerular injury (14,15).
A role of inflammation in the development and DKD progression has been previously proposed (16). In the paper by Fu et al. the authors studied the different types of immune cells that were increased in the glomeruli from diabetic mice. By scRNA-seq the authors found that the predominant immune cells detected were macrophages with high expression of the macrophage markers, namely C1qa, Cd74, and Adgre1. By contrast, only a small number of cells expressed neutrophil markers (6). Macrophages have been previously associated with tissue damage, although they have also a role in host defense and tissue repair (17). As different macrophages phenotypes and functions have been reported, the assignment of M1 and M2 macrophage in the Fu study was performed on the basis of 57 M1 and 33 M2 marker genes, so a cell with higher positive M1 score was considered M1 cell, and a cell with higher positive M2 score was considered M2 cell. As expected, among the identified cluster of macrophages, there was a higher number of cells expressing M1 than cells expressing M2. This shows that inflammatory macrophages are the immune cells often found in glomeruli in DKD (6). In concordance with these results, alternatively activated macrophages also called M2, have shown to exert anti‐inflammatory functions which suggest a potential therapeutic approach with macrophage-based therapy. Guiteras et al. (18) demonstrated that the increase of the ratio of alternative activated macrophages/classically activated macrophages within the diabetic kidney significantly reduced albuminuria and renal fibrosis in DKD probably by modulation of the renal inflammatory response (18). Overall this suggest that the immune response is activated early in diabetic glomeruli and that restauration of a macrophage balance may help to protect against diabetic kidney injury.
The information in the kidney atlas is providing clues for understanding kidney structure and specific cell type expression which helps to understand potential pathogenetic mechanisms involved in different kidney diseases (9). Renal diseases are often cell-type specific. For instance, derangements in specific transport proteins restricted to the alpha intercalated cell in the collecting tubule cause most types of hereditary distal renal tubular acidosis but may also be involved in some acquired forms of this complex tubular disease (19). This concept can be extended to the identification of malfunction genes in cell types that cause glomerular diseases and progression of chronic kidney disease (CKD). The group by Sustzak performed a compartment-based eQTL analysis of human kidney tissue samples using manual microdissection of the two main kidney compartments: glomeruli and tubules. With this approach they identified novel genes and cellular pathways involved in CKD development and subsequently potential new opportunities for its treatment (20). Using single cell analysis technique and mapping the gene expression this group studied genes involved in alterations to identify disease-causing cells types and disease involved pathways. The renal alterations studied were proteinuria, glomerular filtration rate, metabolic acidosis, and hypertension. They found that genes associated with nephrotic range proteinuria were only expressed in the glomerular epithelial cells (podocytes), while decreased glomerular filtration rate were mainly expressed in proximal tubular cells, renal tubular acidosis were only expressed in intercalated cells of the collecting duct, genes associated with serum metabolite levels showed very strong enrichment for proximal tubule, and genes implicated in blood pressure regulation were mostly expressed in collecting duct cells (21). This emerging research approach opens a new window for kidney research and the development of new therapeutic tools directed not to whole kidney regions, but to the single altered kidney cell.
In concordance with other previous reports, Fu et al. also showed by pathway enrichment analysis alterations of angiogenesis and migration pathways in endothelial cells from glomerular diabetic mice (6,22). In addition, in mesangial cells from diabetic glomeruli, pathways of regulation of translation, and protein stabilization were enriched. A limitation of this study is that the analysis of the differentially expressed genes between diabetic and control glomerular cells were only performed in endothelial and mesangial cells. The authors were not able to analyze the differentially expressed genes in the podocytes because a limited number of them could be captured in the diabetic glomeruli (6). This was possibly related to the decreased podocyte number which is characteristic of diabetic kidneys (14,15). Of note, a previous study from Lu et al. examined the single-cell RNA-seq analysis of cultured mouse podocytes and were able to identify 92 genes that were preferentially expressed in and specifically essential for podocytes. Furthermore, they validated the essentiality of these genes for podocytes and identified 30 novel genes that are probably involved in the podocyte cytoskeleton assembly (11). Further studies with single-cell RNA-seq centered in the diabetic and non-diabetic podocyte should help to elucidate the role of this glomerular cell in DKD.
With newly developed techniques whole-genome DNA methylation maps for human kidney samples from healthy and DKD patients differentially methylated regions are being identified. In agreement with the scRNA-seq study in isolated glomerulus, DNA methylation and RNA sequencing demonstrated marked changes in immune cell proportions in kidneys from patients with DKD (23). Differentially methylated regions were highly enriched in genes with metabolic functions and immune response, such as TNF (23). In addition, TNF signaling genes, including TNF expression, were higher and the methylation of the TNF was consistently lower in human samples from DKD as compared with the healthy controls (23). The methylation of the TNF gene locus was consistently lower in diabetic samples (23). With this new strategy one can now detect the expected differences in the methylation status of kidneys from diabetic and non-diabetic patients. Of note, a new generation drugs targeting epigenetic DNA and histone modifications to treat diabetic kidney disease are in the pipeline (24). As the specific single-cell RNA sequencing analyses of isolated kidney cells from normal and pathological tissue expands to various forms of kidney disease a better understanding of kidney disease at the cell-specific level will emerge. This approach should help to develop new diagnostic and therapeutic strategies.
Acknowledgments
Funding: MJ Soler acknowledges grant support from the Fondo de Investigación Sanitaria-FEDER, ISCIII, PI17/00257, and REDINREN, RD16/0009/0030. D Batlle acknowledges grant support from the National Institute of Diabetes and Digestive Kidney Diseases grants U01-DK071733-01A1 and R01DK104785 as well as by a gift to Northwestern University by the Joseph and Bessie Feinberg Foundation. MJ Soler reports consultation fees with NovoNordisk, Janssen, Boehringer, Eli Lilly, AstraZeneca, and Esteve. D Batlle is the founder of Angiotensin Therapeutics Inc. and has received consultant fees from Relypsa Inc.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Woroniecka KI, Park ASD, Mohtat D, et al. Transcriptome analysis of human diabetic kidney disease. Diabetes 2011;60:2354-69. [Crossref] [PubMed]
- Hodgin JB, Nair V, Zhang H, et al. Identification of cross-species shared transcriptional networks of diabetic nephropathy in human and mouse glomeruli. Diabetes 2013;62:299-308. [Crossref] [PubMed]
- Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol Cell 2015;58:598-609. [Crossref] [PubMed]
- Wang X, Park J, Susztak K, et al. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat Commun 2019;10:380. [Crossref] [PubMed]
- Cochain C, Vafadarnejad E, Arampatzi P, et al. Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis. Circ Res 2018;122:1661-74. [Crossref] [PubMed]
- Fu J, Akat KM, Sun Z, et al. Single-Cell RNA Profiling of Glomerular Cells Shows Dynamic Changes in Experimental Diabetic Kidney Disease. J Am Soc Nephrol 2019;30:533-45. [Crossref] [PubMed]
- Soler MJ, Riera M, Batlle D. New experimental models of diabetic nephropathy in mice models of type 2 diabetes: efforts to replicate human nephropathy. Exp Diabetes Res 2012;2012:616313. [Crossref] [PubMed]
- Azushima K, Gurley SB, Coffman TM. Modelling diabetic nephropathy in mice. Nat Rev Nephrol 2018;14:48-56. [Crossref] [PubMed]
- Park J, Shrestha R, Qiu C, et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 2018;360:758-63. [Crossref] [PubMed]
- Ye Y, Song H, Zhang J, et al. Understanding the Biology and Pathogenesis of the Kidney by Single-Cell Transcriptomic Analysis. Kidney Dis (Basel) 2018;4:214-25. [Crossref] [PubMed]
- Lu Y, Ye Y, Bao W, et al. Genome-wide identification of genes essential for podocyte cytoskeletons based on single-cell RNA sequencing. Kidney Int 2017;92:1119-29. [Crossref] [PubMed]
- Rothwell DG, Li Y, Ayub M, et al. Evaluation and validation of a robust single cell RNA-amplification protocol through transcriptional profiling of enriched lung cancer initiating cells. BMC Genomics 2014;15:1129. [Crossref] [PubMed]
- Karaiskos N, Rahmatollahi M, Boltengagen A, et al. A Single-Cell Transcriptome Atlas of the Mouse Glomerulus. J Am Soc Nephrol 2018;29:2060-8. [Crossref] [PubMed]
- Riera M, Márquez E, Clotet S, et al. Effect of insulin on ACE2 activity and kidney function in the non-obese diabetic mouse. PloS One 2014;9:e84683. [Crossref] [PubMed]
- Nadarajah R, Milagres R, Dilauro M, et al. Podocyte-specific overexpression of human angiotensin-converting enzyme 2 attenuates diabetic nephropathy in mice. Kidney Int 2012;82:292-303. [Crossref] [PubMed]
- Navarro-González JF, Mora-Fernández C. The role of inflammatory cytokines in diabetic nephropathy. J Am Soc Nephrol 2008;19:433-42. [Crossref] [PubMed]
- Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 2008;8:958-69. [Crossref] [PubMed]
- Guiteras R, Sola A, Flaquer M, et al. Exploring macrophage cell therapy on Diabetic Kidney Disease. J Cell Mol Med 2019;23:841-51. [Crossref] [PubMed]
- Vallés PG, Batlle D. Hypokalemic Distal Renal Tubular Acidosis. Adv Chronic Kidney Dis 2018;25:303-20. [Crossref] [PubMed]
- Qiu C, Huang S, Park J, et al. Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat Med 2018;24:1721-31. [Crossref] [PubMed]
- Park J, Liu C, Kim J, et al. Understanding the kidney one cell at a time. Kidney Int 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Fu J, Wei C, Zhang W, et al. Gene expression profiles of glomerular endothelial cells support their role in the glomerulopathy of diabetic mice. Kidney Int 2018;94:326-45. [Crossref] [PubMed]
- Park J, Guan Y, Sheng X, et al. Functional methylome analysis of human diabetic kidney disease. JCI Insight 2019.4. [PubMed]
- Fontecha-Barriuso M, Martin-Sanchez D, Ruiz-Andres O, et al. Targeting epigenetic DNA and histone modifications to treat kidney disease. Nephrol Dial Transplant 2018;33:1875-86. [Crossref] [PubMed]


