Applying deep learning in recognizing the femoral nerve block region on ultrasound images
Introduction
Regional nerve block is a common anesthesia technique in recent years. A successful block requires excellent anesthesia experience including the identification of nerves and fascia, and good operative skills. Previous studies have shown that ultrasound-guided technique could significantly increase the success rate of nerve block. However, some studies have found that even the use of ultrasound had a high failure rate, mainly because a number of operators were trainees with less experience and insufficient ultrasonic skills (1). A failed nerve block not only leads to a bad experience during surgery, but also damages the healthy and even threats the life (2). Thus, accurately identifying the nerve block region is important for these operators.
Nowadays, artificial intelligence (AI), especially deep learning (DL) has been applied in all walks of life. Computer vision (CV) is a branch of DL, which plays a crucial role in image processing. Former studies have designed different methods to achieve ultrasound image segmentation, turning out unsatisfied results (3-5). There are few studies applying deep neural network in the femoral nerve segmentation on ultrasound images. In 2016, Kaggle competition held a combat in recognizing the brachial nerve, which inspired us to look for better solutions (6).
In the present study, we adopted a deep neural network well used in biomedicine to train hundreds of ultrasound images of femoral nerve block (7). We aim to construct and share a dataset of ultrasound images of regional nerve block and to explore a method to identify the region of interest for medical images, which may potentially be used in clinical practice.
Methods
Materials preparing and model selecting
We retrospectively collected the ultrasound images of the femoral nerve from the clinic. All images—they were RGB images (it means the image has 3 channels rather than one as the grayscale image) after exported from ultrasound machines—were selected by three experienced physicians to ensure that the femoral nerve regions were clearly identified. In order to increase the heterogeneity of the data, we tried to involve more patients rather than more images from same patients. We removed the peripheral part of each image that did not contain the ultrasound signals. Then we converted the original RGB images to grayscale ones since the ultrasound signal is grayscale. All manual image segmentation was performed by an experienced physician and verified by the other two with the tool of ‘Labelme’ (8). Since the femoral nerve is difficult to identify, we delineated the connective tissue surrounded by the iliofascial membrane and the iliopsoas, the key area for femoral nerve block (9). To ensure anonymity, we confirmed that there was no information on the images to identify the patients. Research Ethics Boards at the First Affiliated Hospital of Sun Yat-sen University approved the protocol. The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
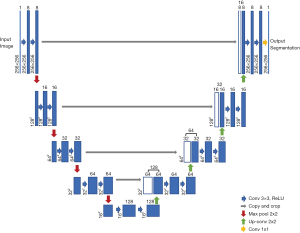
We selected the U-net model to train our data, which is a widely used network for biomedical image segmentation (7). U-net is an upgrading based on fully convolutional network (FCN) (In FCN, all layers are convolutional layers and no fully connected layer exists); its architecture is showed below (Figure 1). The input image is convolved by multiple layers and extracted to numbers of features, which are then deconvolved and add the convolved images in shallow layers. Finally, the image size of the output layer is the same as that of the input layer. The framework is just like the word ‘U’, which is where its name comes from. We fed the ultrasound images to the input layer and set the manually marked masks as the output segmentation map to train the model.
Dataset were randomly divided into the training set, the development set and the test set. Only the training set fitted the model; the development set was used for validation during fitting process; and the test set was used to evaluate the model.
Hyperparameter setting and data preprocessing
Deep neural networks have numbers of hyperparameters. We only focused on the important ones and set the others to default values. To speed up calculations, the input images were resized smaller—256 (width) × 256 (height) × 1 (channels)—as demonstrated earlier by the Demo of Kaggle competitor Kjetil Åmdal-Sævik (10). All hidden layers and the convolution kernel were shown in Figure 1. The activation function for each hidden layer was rectified linear unit (ReLU); for output layer it was sigmoid function. Each pixel of every image was normalized according to Eq. [1], where pn was the normalized pixel while p was the original pixel.

We selected Adam algorithm with default hyperparameters as the optimizer (11). We set batch size as 32 and training epochs as 75 without early stopping.
Measurements definition
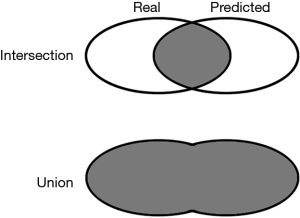
The segmentation quality was evaluated by the metrics of intersection over union (IoU), which was defined as Eq. [2]. The intersection refers to the area of overlap between the predicted mask and manual mask, while the union refers to the combination of these two masks (Figure 2).

Firstly, we calculated the sum IoU (sum intersection over sum union) of all images in training set and test set respectively. Then, each IoU of every image was calculated to measure the precision. An IoU >0.5 was considered as effective segmentation, according which the accuracy was defined as Eq. [3]:
Then, the predicted masks of test set were drawn on the original image to give an intuitive evaluation and comparison with manual segmentation.
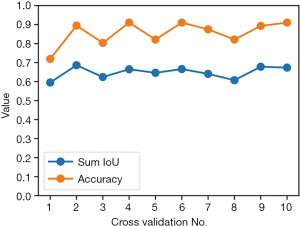
Finally, a 10-fold cross validation was applied to evaluate the robust of results. The whole dataset was enrolled in this section. For each fold, the sum IoU and accuracy were calculated and illustrated.
Analysis tools and platform
Images, also called data in our study, were managed in a sequence of preprocessing, training, prediction and evaluation. The data flow pipeline was compiled by Keras with backend of Tensorflow (12,13), running on Google Colaboratory driven by the GPU of Tesla K80. All processings were coded by python 3.6 with necessary modules. For reproducibility, we have shared our data and codes on Github (https://github.com/gscfwid/femoral_nerve_block_computer_vision).
Results
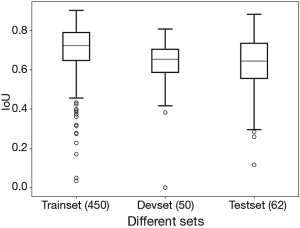
We selected 562 images from 673 ultrasonic images. Among these images, 50 images were randomly selected as the development set while 62 as the test set. After training, the sum IoU was 0.713 for training set and 0.633 for the development set. After predicting, the sum IoU of the test set was 0.638. The distribution of individual IoU of every image in different sets was displayed in Figure 3. The median IoU and upper/lower quartiles were 0.722 (0.647–0.789), 0.653 (0.586–0.703), 0.644 (0.555–0.735) for the train set, development set and test set respectively. The segmentation accuracy of the test set was 83.9%.
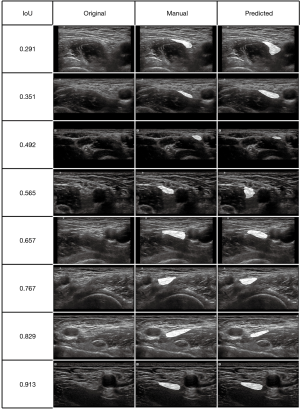
We selected some representative predicted masks of the test set from different IoU levels and highlighted them on the original image (Figure 4).
The 10-fold cross validation was performed on the all 562 images; one tenth of them were for validation and the rest were for training. After fitting and predicting, the accuracies and sum IoUs of validation set for the ten iterations were calculated and illustrated on Figure 5. The median and quartiles of the 10-iteration sum IoUs were 0.656 (0.628–0.672); for accuracy, they were 88.4% (82.1–90.7%).
Discussion
Semantic segmentation is a subtype of CV. U-net is a powerful tool for image segmentation. Recently, the U-net model has been widely adopted in more and more medical image processing (14-16). Similar with FCN, both the input and output of U-net are images. The deeper layers have more abstract characteristics while the shallower layers contain more location details (17). The U-net model up-samples the deeper layers and concatenates with the shallow layer to obtain both target characteristics and location information. This is the basic theory of U-net image segmentation.
In our dataset, images were acquired by ultrasonic machines with different manufacturers (e.g., SonoSite, Wisonic). The ultrasound transducers, focal depths and scan modes during image acquisition were also different. As shown in Figure 4, images with worse IoUs were characterized by larger field of vision and deeper depth (like the second and third rows); mainly because there were relatively less images with characteristics above in the training set. If a larger training dataset with more comprehensive data is available, we will get better segmentation.
At present, CV is widely used in the diagnosis and localization of tumors. Its clinical application for regional nerve block is insufficient, despite we are not the very first to apply this technique in nerve segmentation. Smistad et al. and his fellows have tried the U-net into highlighting the nerves and blood vessels for ultrasound-guided axillary nerve block (18). But it was limited by the very small dataset (only 49 subjects). Zhao and Sun improved the U-net in the ultrasonic images of brachial plexus block and promoted the performance of segmentation (19). The studies above demonstrated a great potential for U-net to apply in nerve segmentation. Besides nerve, U-net can identify other tissues (nerve, blood, muscle, etc.) as long as enough corresponding images could be marked and learned. It can also run very fast and be good at real-time segmentation, which may be introduced as basic functions of ultrasonic machine in the future. Notably, the data labeling is a time-consuming work that requires professional medical experience, which brings difficulties for its application in the medical fields.
In the present study, we introduced a U-net model to solve the ultrasound image segmentation. Despite a relatively small dataset was used for training, the performance of the U-net model is satisfactory finally. However, our study still has several shortcomings. First, the dataset was so small, lacking images with large field of vision and deep depth. Second, we didn’t use data augmentation, which may help to improve the image segmentation capabilities. In addition, we only used a simplified U-net to train the data. There may be a slight discount in performance despite the faster training and forecasting. After further improving, we believe that the model will have great clinical application potential.
Conclusions
We provided a dataset and trained a model for femoral-nerve region segmentation with U-net, obtaining a satisfactory performance. This dataset can help train and evaluate other similar models in the future. The image-segmentation technique may have potential clinical application.
Acknowledgments
We appreciate such an open environment in the field of AI. We thank everyone for their contributions to the computer languages, modules, models, and frameworks we use.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Research Ethics Boards at the First Affiliated Hospital of Sun Yat-sen University approved this study ([2019] 368).
References
- Abrahams MS, Aziz MF, Fu RF, et al. Ultrasound guidance compared with electrical neurostimulation for peripheral nerve block: a systematic review and meta-analysis of randomized controlled trials. Br J Anaesth 2009;102:408-17. [Crossref] [PubMed]
- Baby M, Jereesh AS. Automatic nerve segmentation of ultrasound images. 2017 International conference of Electronics, Communication and Aerospace Technology (ICECA). Available online: https://ieeexplore.ieee.org/document/8203654
- Hadjerci O, Hafiane A, Conte D, et al. Computer-aided detection system for nerve identification using ultrasound images: A comparative study. Informatics in Med Unlocked 2016;3:29-43. [Crossref]
- Hadjerci O, Hafiane A, Makris P, et al. Nerve Detection in Ultrasound Images Using Median Gabor Binary Pattern. In: Campilho A, Kamel M. editors. Image Analysis and Recognition. Springer International Publishing, 2014:132-40.
- Hadjerci O, Hafiane A, Makris P, et al. Nerve Localization by Machine Learning Framework with New Feature Selection Algorithm. In: Murino V, Puppo E. eidtors. Image Analysis and Processing—ICIAP 2015. Springer International Publishing, 2015:246-56.
- Kaggle. Ultrasound Nerve Segmentation. Available online: https://www.kaggle.com/c/ultrasound-nerve-segmentation
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells WM, et al. editors. Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015. Springer International Publishing, 2015:234-41.
- Russell BC, Torralba A, Murphy KP, et al. LabelMe: A Database and Web-Based Tool for Image Annotation. Int J Comput Vis 2008;77:157-73. [Crossref]
- Jeyaraj SK, Pepall T. Ultrasound guided femoral nerve block anaesthesia tutorial of the week 284. Available online: https://www.wfsahq.org/components/com_virtual_library/media/7e454e1aacaa79db571a422a5ac6354f-284-Ultrasound-Guided-Femoral-Nerve-Block.pdf
- Åmdal-Sævik K. Keras U-Net starter-LB 0.277. Kaggle. Available online: https://www.kaggle.com/keegil/keras-u-net-starter-lb-0-277
- Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. 2014. arXiv:1412.6980.
- Abadi M, Barham P, Chen J, et al. TensorFlow: A System for Large-Scale Machine Learning. OSDI 2016. Available online: https://www.usenix.org/system/files/conference/osdi16/osdi16-abadi.pdf
- Keras: The Python Deep Learning library. Astrophysics Source Code Library. 2018. Available online: https://ascl.net/1806.022
- Christ PF, Ettlinger F, Grün F, et al. Automatic Liver and Tumor Segmentation of CT and MRI Volumes using Cascaded Fully Convolutional Neural Networks. arXiv:1702.05970.
- Dalmış MU, Litjens G, Holland K, et al. Using deep learning to segment breast and fibroglandular tissue in MRI volumes. Med Phys 2017;44:533-46. [Crossref] [PubMed]
- Dong H, Yang G, Liu F, et al. Automatic Brain Tumor Detection and Segmentation Using U-Net Based Fully Convolutional Networks. Medical Image Understanding and Analysis. 2017. arXiv:1705.03820.
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Smistad E, Johansen KF, Iversen DH, et al. Highlighting nerves and blood vessels for ultrasound-guided axillary nerve block procedures using neural networks. J Med Imaging (Bellingham) 2018;5:044004. [Crossref] [PubMed]
- Zhao H, Sun N. Improved U-Net Model for Nerve Segmentation. In: Zhao Y, Kong X, Taubman D. editors. Image and Graphics. Springer International Publishing, 2017;496-504.