
microRNA-1 inhibits cardiomyocyte proliferation in mouse neonatal hearts by repressing CCND1 expression
Introduction
Previous studies have shown that urodele amphibians and zebrafish have extraordinary abilities to regenerate their heart following injuries (1,2). In adult zebrafish, they can regenerate their heart after 20% of the ventricle was removed. This was achieved without fibrotic scar formation and mainly through cardiomyocyte proliferation. However, this remarkable ability is not seen in mammalian hearts. It has been reported that cardiomyocytes in 2 day-old mouse neonates could still divide extensively in all regions of the heart (3,4). Unfortunately, this proliferative ability is greatly reduced by the time that the neonatal heart reaches 13 days old. In the adult mammalian heart, the cardiomyocytes are “irreversibly” growth arrested so are unable to self-renewal following myocardial damage. Surprisingly, Porrello et al. (5) reported that 1-day-old mouse neonates could completely regenerate their myocardium after 15% of their left ventricle was removed. Detailed analyses of the results revealed that the regeneration was achieved through the proliferation of pre-existing cardiomyocytes, while the architecture of the heart and physiological functions were also restored. However, 7-day-old neonates lose this regenerative ability and fibrotic scars replace the missing myocardium. In this context, it is important to elucidate the underlying mechanisms that regulate neonatal cardiomyocyte proliferation, which could potentially be translated in regenerative medicine to treat degenerative heart diseases.
Eulalio et al. (6) have reported that two miRNAs (hsa-miR-590 and hsa-miR-199a) possess the ability to facilitate adult cardiomyocytes’ re-entry into the cell cycle ex vivo. In addition, following myocardial infarction in mice, expression of these miRNAs could be used as a marker to assess the extent of myocardial recovery. It has been reported that miR-25 could potently delay calcium influx in cardiomyocytes in vitro and increased the risk of heart failure in humans and mice (7). Furthermore, injection of antagomiR against miR-25 could prevent heart failure in mice by improving cardiac function and cardiomyocyte survival. It now appears that altered patterns of miRNAs expression are not only involved in a variety of cardiac disorders (8) but also demonstrated promising results in enhancing cardiac regeneration.
miRs belong to a class of single strand nucleotides, which are about 22 nt long and are composed of highly conserved small non-coding RNAs that regulate gene expression at post-transcriptional level (9). Mature miRNAs function by binding to the 3’-untranslated region of target genes that contain the complementary base-paring. The pairing process results in transcriptional repression or degradation of the target mRNAs, which may be involved in organ development, cell proliferation, differentiation and apoptosis—mainly through the negative regulation of protein expression (10). miRs’ sequences are highly conservative in different species, demonstrating tissue-specific and developmental stage-specific expressions. They are adept at regulating the expression of transcription factors and signaling mediators, which play imperative roles in the cardiac development processes and proper timely functions. Presently, we investigated and identified miRNAs distinctly expressed in 2-day-old and 13-day-old mice hearts. Furthermore, we also examined the relationship between these differentially expressed miRs and their target mRNAs with respect to cardiomyocytes proliferation.
Methods
Animals
Two- and 13-day-old ICR neonates were obtained from Laboratory Animal Services Centre, Chinese University of Hong Kong. All experimental protocol used in this study was approved by the Chinese University of Hong Kong Animal Care and User Committee and Department of Health (Ref No.: 13-2 in DH/HA&P/8/2/1 Pt.28).
Primary cardiomyocyte cultures
Primary cardiomyocyte cultures were established from 2-day-old mouse neonate hearts. Briefly, the hearts were isolated from the mice, dissected, rinsed and minced in HEPES-buffered saline solution (HBSS). The tissues were then enzymatic dissociated using 0.1% collagenase I in HBSS at 37 °C for 1 hr. The dissociated cell suspension was centrifuged at 1,000 rpm for 5 min and resuspended in DMEM/ F12, (Invitrogen) containing 10% fetal bovine serum, 100 units/ml penicillin and 100 mg/mL streptomycin. Subsequently, the cells were maintained in a humidified incubator at 37 °C and 5% CO2.
Tissue cryo-sectioning and immunofluorescent staining
Briefly, 2- and 13-day-old mice hearts were freshly harvested, fixed in 4% paraformaldehyde and dehydrated in 30% sucrose solution overnight (11). The hearts were then embedded in OCT and sectioned at 5 µm on a Cryotome (ThermoFisher Scientific, USA). The sections were mounted onto glass slides and then washed twice in PBS for 10 minutes. For immunofluorescence staining, the sections were first permeabilized in 0.3% Triton X-100 and 0.1% SDS in PBS solution for 30 min, followed by blocking in 2% BSA plus 5% donkey serum for 1 hr. The sections were then stained overnight at 4 °C with the following primary antibodies (1:200) diluted in blocking solution: mouse anti-cardiac troponin T antibody (cTnT, Abcam), Rabbit Ki-67 antibody (Abcam) and Rabbit pH3 antibody (Santa Cruz Biotechnology). The tissues were washed three times with PBST (PBS buffer with 0.05% Tween 20) for 15 min to remove unbound primary antibodies. Then the appropriate secondary antibody (1:300) conjugated to Alexa Fluor-488, -555 or -647 (Life Technologies) was added for 1 hr at room temperature, in the dark with gentle shaking. The unbound secondary antibody was removed by washing with PBST, three times for 10 min and PBS for 5 min. The nuclei were counter-stained with 4, 6-diamidino-2-phenylindole (DAPI, Molecular Probes) in 70% (v/v) glycerol and mounted with glass cover slips. Immunofluorescence images were captured under a confocal microscope (FV1000; Olympus, Japan) with fixed exposure settings for all of the samples (12).
miR-1 mimics and antagomir functional analysis
miR-1 mimics and antagomir were purchased from GenePharmaTM (Shanghai, China) and used for the functional analysis. microRNA sequences were generated from Caenorhabdities elegans and served as a negative control (N.C.) in the mimics and antagomir experiments. The sequences are listed in Table S1. The cell transfection experiments were performed using Lipofectamine® RNAiMAX Reagent (Invitrogen Life Technologies, USA) and instructions according to the manufacturer. Briefly, 3.5×105 cardiomyocytes were seeded into each well of a 6-well culture plate. These cells were then transfected with miR-1 mimics, mimics N.C. (control), anti-miR-1 and anti-miR N.C. (control), using Lipofectamine RNAiMAX.

Full table
Stem-loop quantitative real-time miRNA assay
Total RNA, including microRNA, were extracted from heart tissue samples or cultured cardiomyocytes using a NucleoSpin Total RNA Isolation Kit (Clontech Laboratories, USA), according to the manufacturer’s instruction. After DNase I (Takara, Japan) digestion for 15 min, the RNA concentration was determined using a NanoDrop™ 2000 Spectrophotometer (ThermoFisher Scientific, USA). Realtime miRNA assay was performed in two steps: stem-loop RT reaction and quantitative realtime (qRT) PCR detection. RNA was reverse-transcribed into cDNA with specific RT primers (Table S1) by using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA). RT-qPCR was then performed to assay miRNA expression using the specific forward primers and the universal reverse primer complementary to the anchor primer. PCRs were conducted in triplicates using an ABI 7900HT Real-Time PCR Detection System (Applied Biosystems, USA) and 2× SYBR Premix Ex Taq (Takara, Japan) at 95 °C for 30 s, 40 cycles of 95 °C for 5 s and 60 °C for 30 s. After RT-qPCR amplification, a dissociation analysis (melt curve) was performed to identify the characteristic peaks associated with primer-dimers in order to separate them from the single prominent peak that represents the successful PCR amplification of miRs. Relative expression level of a given miRNA was determined by delta-delta (∆∆) Ct calculation and reference to normalized to U6 snRNA control level. Expression levels of miRs were presented as fold change (2-∆∆Ct). The data was expressed as mean and standard deviation of relative values obtained from three different samples. T-tests were performed with significant p-value set at 0.05 for comparison. The PCR primers for mature miR-1 or U6 were designed as follows: miR-1 forward primer, 5'-CGGCGGTGGAATGTAAAGAAG-3' and miR-1 reverse primer 5'-GTGCAGGGTCCGAGGT-3'. U6 snRNA forward primer, 5'-TTCCTTGGCCCCTGCGCA-3' and reverse primer, 5'-GTGCAGGGTCCG AGGT-3'. Other miRs primer sequences are reported in Table S1.
RT-qPCR assays
Total RNA was extracted from cultured cardiomyocytes and heart tissue samples as described above. One µg of total RNA was reverse-transcribed into cDNA with oligo (dT18) primer using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA) according to the manufacturer’s instruction. Briefly, 20 µL of the reactant was incubated for 60 min at 42 °C, 5 min at 70 °C, and then stored at −20 °C. A SYBR Green Master Mix Kit was used in the qRT–PCR to quantify expression of the target genes. GAPDH mRNA levels were used for normalization. The analysis methods used was similar to that mentioned in the Stem-loop Quantitative Realtime miRNA Assay. The primer sequences used analyzing murine FGF4, Gas2l1, IGF-1, Frs2, FoxP1 Cdc14a, CCND2, CCND1 and CDC14a expression are listed in Table S1.
Cell viability assay
MTT assays were performed to establish the viability of neonatal cardiomyocytes after miR-1 mimics and antagomir treatments. Briefly, 200 µL of 1×104 neonatal cardiomocytes was seeded into each well of a 96-well plate. These cells were allowed to attach for 24 hr and then transfected with 100 nmol/L miR-1 mimics, mimics negative control (N.C.). After 48 hr of transfection, 20 µL of 5 mg/mL MTT (Sigma, USA) was added to the cultures and incubated for another 4 hr at 37 °C and 5% CO2. The MTT solution was then removed and the precipitated formazan crystals were dissolved in 200 µL DMSO. The optical density (OD) value for the dissolved formazan was measured on a microplate reader (Bio-Rad) with wavelength set at 490 nm.
BrdU cell proliferation analysis
The effect of miR-1 on cardiomyocytes proliferation was investigated using Bromodeoxyuridine (BrdU) incorporation analysis. Briefly, cardiomyocytes were transfected with 100 nmol/L miR-1 mimics or negative mimic control and maintained in growth medium for 48 hr. The cells were then pulse labeled for 4 hr in the presence of 10 µM of BrdU introduced into the culture medium. After pulse labelling, the cells were fixed and processed for immunoflorescent staining using mouse monoclonal BrdU antibody (1:200, Invitrogen). The BrdU incorporation rate was determined as the percentage of BrdU+ cells over the total number of DAPI+ cells in each randomly selected microscopic field. On average, 10 microscopic fields from three independent experiments were evaluated.
Flow cytometry analysis
The cell cycles of control and experimental cardiomyocytes were assessed and determined using flow cytometry. These cells were plated into 6-well plates at 3.5×105 cells per well and allowed to grow for 24 hr. The cells were transfected with 100 nmol/L miR-1 mimics, negative mimic control (N.C.), miR-1 antagomir and antagomir N.C. for 48 hr using Lipofectamine RNAiMAX Reagent (Invitrogen,CA). The cells were then trypsinized, collected by centrifugation, washed in cold PBS and fixed overnight at 4 °C in 70% ethanol/PBS. Subsequently, the cells were centrifuged and washed twice with PBS and stained with 20 µg/mL propidium iodide (Sigma-Aldrich, USA), 100 µg/mL RNase (Roche) and 0.1% Triton-X100 to a final volume of 200 µL at room temperature for 30 min. The cell cycle was analyzed using a flow cytometer (FACS Calibur, Becton Dickinson, USA) at 542 nm. The experiment was conducted three times and at least 10,000 events were analyzed for each sample. The cell phase percentages were processed and determined using a Modfit software.
Immunocytofluorescent staining
Cardiomyocytes (0.75×104 per well) were seeded onto ϕ1.2 cm glass coverslips (Thermo Scientific, USA) in a 24-well tissue culture plate for 24 hr. The cells were transfected with 100 nmol/L miR-1 mimics and N.C. for 48 hr as described above. Subsequently the samples were washed twice with PBS and fixed in 4% paraformaldehyde for 30 min. The cells were then permeabilized with 0.3% Triton X-100 and 0.1% SDS in PBS solution for 30 min, followed by blocking in 2% BSA with 5% donkey serum for 1 hr. These cells were stained overnight at 4 °C with the following primary antibodies (1:200 dilution in blocking solutions): Mouse Anti-Cardiac Troponin T antibody (Abcam), Rabbit Anti-IGF1 antibody (Abcam), Rabbit pH3 antibody (Santa Cruz Biotechnology) and Rabbit Anti-CCND1 antibody. The specimens were then washed three times with PBST (PBS buffer with 0.05% Tween 20) for 15 min to remove unbound primary antibody. Then treated with the appropriate secondary antibody (1:300 Abcam) conjugated to Alexa Fluor-488, -555 or -647 (Life Technologies) was added for 1 hr at room temperature in the dark on a shaker. All unbound secondary antibodies were removed by washing with PBST (3X) for 10 min and PBS for 5 min. The nuclei were counterstained with DAPI (Molecular Probes, USA) in 50% (v/v) glycerol and mounted onto the glass slides. The stained specimens were analyzed and recorded on a confocal microscope (FV1000; Olympus, Japan).
Predication of target genes regulated by miR-1
miRNAs function by binding to the 3’UTR of their target mRNAs which causes the mRNA to degrade or represses their translation. We have searched the potential target genes of miR-1 using online softwares and databases (TargetScan, PicTar, miRBase and miRanda). We found that the mature sequence of miR-1 was highly conserved between different species, including mice, rat and human. Among the 695 predicted conserved targets, we highlighted FGF4, Gas2l1, IGF-1, Frs2, FoxP1 Cdc14a, CCND2, CCND1 and CDC14a, which playing important roles cell proliferation and differentiation.
Construction of luciferase reporters
We have constructed vectors that included the 3’UTR binding site of FGF4, Frs2, FoxP1 or CCND1 mRNAs to examine whether miR-1 could directly bind these 4 mRNAs in cardiomyocytes. The 3’UTR segments (400–600 bp) of target mRNAs, predicted to interact with miR-1, were amplified by PCR from cDNA that was reversely transcribed from RNA extracted from the 2-day-old hearts. The PCR primers used during amplification are listed in Table S1. The amplified fragments were individually subcloned upstream of the luciferase coding sequence of Firefly/Renilla dual reporter vector pmirGLO (Promega, Madison, USA). The correct insertions of vectors were confirmed by sequencing (BGI, China).
Dual-glo luciferase reporter assays
The direct interaction between miR-1 and 3’UTR of FGF4, Frs2, FoxP1 or CCND1 mRNAs were assessed. Neonatal cardiomyocytes were transiently co-transfected with 100 ng recombinant luciferase vectors (containing IGF1, Frs2, FoxP1, CDK9 or CCND1-3’UTR) and 100 nM miR-1 mimics or mimics N.C. using Lipofectamine 2000 (Invitrogen). At 48 hr transfection, luciferase activity was measured using the Dual-Glo luciferase assay system (Promega, Medison). The firefly luciferase activity was normalized against renilla luciferase for each individual analysis. The values in the miR-1 mimics group were compared with those transfected with mimics N.C. and pmirGLO empty vector. The results were presented as mean ± standard deviation (SD) from triplicated experiments.
Western blot analysis
Cardiomyocytes transfected with 100 nmol/L miR-1 mimics or negative control were washed twice with cold PBS and lysed on ice in 200 µL RIPA buffer (50 mM sodium chloride, 1% Nonidet P-40, 20 mM Tris, pH 7.6) in the presence of protease inhibitor cocktail (Complete ULTRA Tablets, Roche Applied Science, USA) and 100 µg/mL PMSF for 30 min. The cell lysates were then centrifuged at 12,000 rpm at 4 °C for 20 min. The protein supernatant was transferred into Eppendorf tubes and stored at −80 °C. The protein concentration of each sample was measured using a Bio-Rad protein assay kit (Bio-Rad, USA). Equal amount (50 µg) of cell lysates was loaded onto 10% SDS-PAGE and run for 2 hr at constant voltage (stacking gel: 80V, 30 min; separating gel: 200V, 90 min). A Trans-Blot semi-dry electrophoretic transfer cell (Bio-Rad) was used to electrophoretically transfer the separated proteins onto PDVF membranes (GE Healthcare), which were then blocked with 5% skimmed milk in TBST (10 mM Tris pH 7.6, 50 mM NaCl, 0.05% Tween-20) for 1 hr at room temperature. The membranes were first incubated with 1:1,000 diluted primary antibodies (anti-CCND1, anti-IGF1, anti-Tubulin β3) at 4 °C overnight. The blots were then washed with TBST (TBS buffer with 0.05% Tween 20), then probed with IRDye 800CW-labeled secondary antibodies and IRDye 680LT-labeled secondary antibodies (Li-COR, USA). After washing, the images were viewed and recorded using an Odyssey scanning system (Li-COR, USA).
Statistical analysis
All data presented were generated from triplicate samples. Results were calculated using a GraphPad Prism computer software and reported as mean ± SD. Comparison of differences between groups was determined by Student’s t-test and analysis of variance (ANOVA). Statistical significance was defined as P<0.05.
Results
miRNAs differentially-expressed in 2- and 13-day-old mice hearts
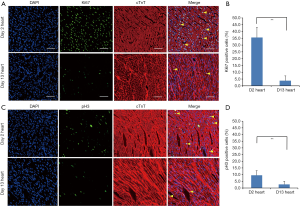
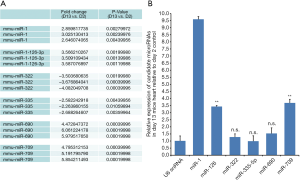
Using immunofluorescent staining for proliferation marker Ki67, we have demonstrated that the cardiomyocytes actively proliferated in 2-day-old neonate hearts but this rapidly ceases at 13 days. We estimated that 35.6%±7.4% of 2-day-old cardiomyocytes were Ki-67+ verses 3.8%±3.5% in 13-day-old cardiomyocytes (Figures 1A,B). We also determined that the percentage of pH3+ cardiomyocytes was significantly decreased at day 13 (9.5%±3.4% vs. 2.7%±2.2% at day 13 vs. day 2, Figures 1C,D). In this context, we examined the involvement of miRNAs in cardiac proliferation and differentiation. The global miRNA profiling of 2- and 13-day-old mice hearts were established and analyzed (http://fp.amegroups.cn/cms/atm.2019.08.68-1.pdf). The microarray identified six miRNAs that were differentially expressed (Figure 2A). We use RT-qPCR to validate the expression of these five miRNAs and determined that miR-1, miR-126 and miR-709 were significantly up-regulated in 13-day-old hearts compared with 2-day-old hearts. However, miR-322, miR-335 and miR-690 were not differentially expressed between the two time points. Moreover, miR-1 is the most significantly up-regulated miRNA in the 13-day-old heart (Figure 2B).
Effects of miR-1 over-expression on cardiomyocyte proliferation and viability
Cardiomyocytes were isolated from 2-day-old neonates and transfected with miR-1 mimics, mimics N.C. (control), miR-1 antagomir and antagomir N.C. (control) The effects of the transfections were examined by flow cytometry. The results revealed that the percentage of cardiomyocytes in G0/G1 phase of the miR-1 mimics group was significantly increased compared with that in mimics N.C. group. This was accompanied by a concomitant decrease of cell number in the S phase. Moreover, the percentage of cardiomyocytes in G0/G1 phase for the miR-1 antagomir group was significantly decreased compared with the antagomir N.C. group (Figures 3A,B). These data suggest that over-expression of miR-1 in 2-day-old cardiomyocytes reduces the number of these cells in S phase but increases them in the G0/G1 phase.
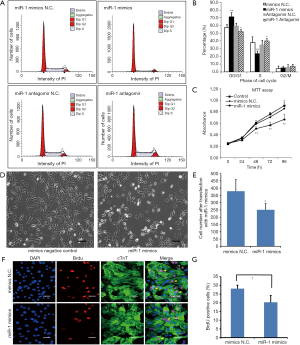
We also investigated the effects of miR-1 on cardiomyocyte viability. MTT assays were performed after the cells were transfected with miR-1 mimics or mimics N.C. These results show that cardiomyocyte viability was significantly decreased following miR-1 mimics compared with mimics N.C. and also without any treatment (Figure 3C, P<0.05). The results suggest that miR-1 reduces cardiomyocyte viability.
The effects of miR-1 on cardiomyocyte proliferation were further investigated by BrdU-incorporation assay. We observed that the cell number was significantly decreased following transfection with miR-1 mimics (Figures 3D,E). Furthermore, the proportion of BrdU+ cardiomyocytes was significantly decreased compared with cardiomyocytes transfected with mimics N.C. (Figures 3F,G). These results indicate that over-expression of miR-1 inhibits cardiomyocyte proliferation.
Predicated target genes regulated by miR-1
In order to understand how miR-1 inhibited cardiomyocyte proliferation, we searched available databases for potential target genes relevant to the biological process using computational online software. We found that miR-1 could potentially bind with the 3'UTR sites of FGF4, Gas2L1, IGF-1, Frs2, FoxP1 Cdc14a, CCND2, CCND1 and CDC14a mRNAs, which contains highly conserved miR-1 seed-matching sequences. FGF-4, IGF-1, CCND1 and Cdc14a encode the for peptides: fibroblast growth factor 4, insulin-like growth factor 1, cyclinD1 and CDC14 cell division cycle 14 homolog A, respectively. We determined that Frs2, FoxP1, IGF-1 and CCND1 expressions were significantly down-regulated compared with expression of these genes in cardiomyocytes transfected with negative control oligonucleotides at 48 hr after miR-1 mimics were transfected into 2-day-old cardiomyocytes (Figure 4A).
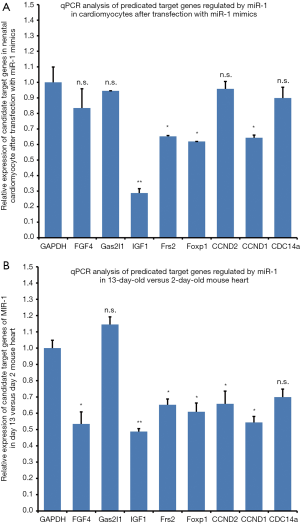
The question that we wanted address was whether the predicated target genes of miR-1 were also down-regulated in the 13-day-old cardiomyocytes (developmental stage when cardiomyocytes have almost stopped dividing). We used RT-qPCR to establish the mRNA abundance of these genes in 2-day-old and 13-day-old mice hearts to elucidate whether there was a correlation between the expressions of these genes in vivo with our in vitro results. We established that FGF4, IGF-1, Frs2, FoxP1, CCND1 and CDC14a expressions were significantly down-regulated in the 13-day-old heart compared with 2-day-old mouse heart (Figure 4B). These results suggest that the rapid decline in cardiomyocyte proliferation during the post-natal period is associated with a corresponding decrease in miR-1 targeted genes expression (genes that are associated with the cell cycle). Furthermore, IGF-1, Frs2, FoxP1 and CCND1 expression were down-regulated in both miR-1 transfected neonatal and adult cardiomyocytes.
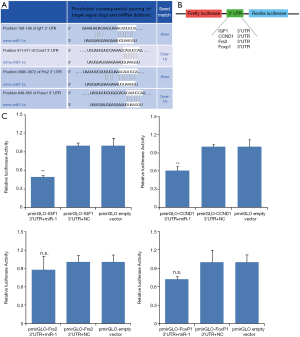
Identification of mRNA Targets for miR-1
We attempted to confirm that IGF-1, Frs2, FoxP1 and CCND1 3’UTR (Figure 5A) were direct targets of miR-1. pmirGLO-UTR vectors (Figure 5B) containing the miR-1 binding sites were constructed to perform reporter assays and the pmirGLO empty vector without 3'UTR was set as a control. All the four constructs were validated by restriction enzyme digestion and sequencing. The luciferase activity of the reporter vectors was assayed 48 hr after co-transfection of the reporter construct with miR-1 mimics or negative control in 2-day-old cardiomyocytes. Following the co-transfection of neonatal cardiomyocytes with miR-1 mimics, the relative luciferase activity of pmirGLO-IGF1 3’UTR and pmirGLO-CCND1 3'UTR were significantly suppressed by 52%, respectively (Figure 5C) in comparison to the co-transfection with the negative control. In addition, the relative luciferase activity did not change when co-transfected with miR-1 and pmirGLO empty vector, indicating that IGF1 3'UTR and CCND1 3'UTR were direct targets of miR-1.
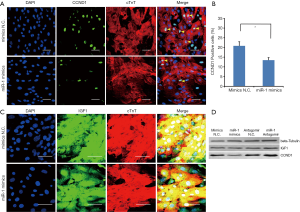
IGF-1 and CCND1 expression in cardiomyocytes over-expressing miR-1
Since miR-1 can inhibit G1/S transition, we decided to examine the effects of miR-1 over-expression on CCND1 and IGF1 which regulate cardiac cell cycle and growth respectively. Using immunofluorescent staining, we examined IGF-1 and CCND1 expression in 2-day-old cardiomyocytes 48 hr after miR-1 mimics or mimics N.C. transfection. Immunofluorescence staining showed that CCND1 expression was repressed in cardiomyocytes transfected with miR-1 mimics (Figures 6A,B). However, IGF-1 expression was unaffected when cardiomyocytes were transfected with miR-1 mimics (Figure 6C). Western blotting was also performed and the results revealed that CCND1 expression was significantly down-regulated in cardiomyocytes 48 hr after miR-1 mimics transfection as compared with miR-1 mimics compared with miR-1 mimics N.C. (negative control). However, IGF-1 expression was unaffected by the miR-1 mimics (Figure 6D). The data suggests that over-expressing miR-1 mimics represses CCND1 expression.
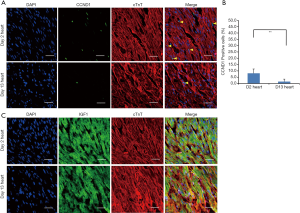
IGF-1 and CCND1 expression in 2- and 13-day-old hearts
It has been reported that IGF1 is an important cytokine for cell cycle progression and growth. In cardiomyocytes, it regulates cell proliferation, apoptosis and other biological processes. CCND1 is a key regulator of cell cycle, so we decided to examine IGF1 and CCND1 expression in 2- and 13-day-old hearts. We determined that CCND1 expression was down-regulated in 13- verses 2-day-old hearts (Figure 7A,B). However, the immunofluorescent staining did not show any obvious difference in IGF-1 expression between the two time points (Figure 7C).
Bioinformatics analysis of differentially expressed mRNA and mRNA targets of miR-1
We have previously reported that the transcripts most significantly down-regulated in 13-day-old cardiomyocytes, were associated with the “cell cycle” as revealed by bioinformatics analysis (13). In addition, above we have demonstrated that is miR-1 was down-regulated in 13-day-old hearts (Figure 2B). These findings allowed us to narrow down our search for differentially expressed mRNA and the predicated mRNA targets of miR-1 that were associated with cardiomyocyte proliferation. Comparison of the two lists for overlapping genes revealed that IGF1 and CCND1 transcripts were down-regulated and simultaneously are amongst the predicated mRNA targets of miR-1 (Table S2). Bioinformatics analysis of the differentially expressed mRNA (13) and mRNA targets of miR-1 (http://fp.amegroups.cn/cms/atm.2019.08.68-2.pdf) further emphasized that IGF1 and CCND1 were targets of miR-1 in neonatal cardiomyocyte.

Full table
Discussion
Cardiomyocyte growth and differentiation are precisely regulated and involves the cell cycle. The different phases of the cell-cycle are orchestrated by positive and negative regulatory proteins. Hence, it is important to elucidate the proteins and miRNAs that control the cardiomyocyte cell-cycle progression and arrest. It will potentially provide tools for resetting the growth-arrested adult cardiomyocytes back into proliferation and repair degenerative heart conditions. It is generally thought that adult cardiomyocytes in mammals are incapable of self-renewable, as these cells are growth arrested shortly after birth. Neonatal cardiomyocytes will stop dividing at around 7 days after birth in mice and 3–4 days in rats (5). In these hearts, 85% of the cardiomyocytes are arrested in G0/G1 phase as determined with flow cytometry. For humans, cardiomyocytes proliferation begins to stop at 3–6 month after birth, with the cells arrested in the G0/G1 phase (14-16). The G0/G1 phase of cell cycle is regulated by cyclins that include cyclin C, D and E. Cyclin D1 plays an important role in G1/S phase transition and proliferation of cells (17) but it could not escape from being down-regulated in the mouse heart after the first week of birth (18).
Cardiovascular disease is one of the leading causes for death in both developed and developing countries (18). Abnormal expression of key genes and signaling transduction are involved in the pathogenesis of cardiovascular diseases. Amongst this, miRNAs are newly found molecules that regulate about 30% of the total gene expression in the human genome, at the post-transcriptional level (19). Numerous studies have demonstrated that miRNAs play an important role apoptosis, cell growth and differentiation, implying that miRNAs are important regulators of development and physiological processes. Hence, miRNAs dysfunction and their roles in fine tuning of mRNA translation may pathologically lead to various diseases (20). Tissue-specific expression is an important feature of miRNAs and miR1, miR126-3P, miR-133, miR499 and Let-7 are abundantly expressed in the heart (21,22). It has been reported that miR-1 is specifically expressed in cardiac and skeletal muscle tissues (23-26). In this study, we have validated that miR-1 is expressed more intensely in the 13- than 2-day-old hearts. In 13-day-old hearts, most of the cardiomyocytes have stopped dividing and have entered into “irreversible” growth arrest.
We examined cardiomyocyte viability, cell cycle progression and BrdU incorporation following miR-1 mimics over-expression in 2-day-old cardiomyocytes. MTT assay was used to assess cell viability. The assay is based on succinate dehydrogenase present in mitochondria of living cells that can reduce exogenous MTT to Formazan. Cells that are dead do not have this function so will not produce Formazan. Our results revealed that cardiomyocyte viability was significantly decreased following miR-1 mimics transfection. Our flow cytometry analysis, also indicated that miR-1 increased the number of cardiomyocytes in the G0/G1 phase but decreased the number of cells in S phase. This implies that these cardiomyocytes were blocked at G1/S transition. In support, the proportion of BrdU+ cardiomyocytes in culture was significantly decreased following transfection with miR-1 as compared with mimics N.C (control). We concluded that miR-1 inhibited cardiomyocyte viability, G1/S cell cycle transition, and proliferation.
We predicted that IGF1 and CCND1 were candidate target genes regulated by miR-1. In addition, our RT-qPCR and the luciferase reporter assays demonstrated that miR-1 could suppress the expression of these two genes at the mRNA level. Our immunocytofluorescent staining and western blot results also confirmed that miR-1 inhibited CCND1 expression at the protein level. In support, we have shown that miR-1 was strongly expressed in the 13-day-old (growth arrested) heart, while CCND1 expression was correspondingly downregulated when compared with 2-day-old (proliferative) heart.
This study has identified miR-1 as an effective inhibitor of CCND1 expression during cardiomyocyte proliferation in the mouse neonatal heart. We have determined that miR-1 could regulate IGF1 expression at the mRNA level but could not demonstrate a corresponding change at the protein level, when cardiomyocytes were transfected with the miR-1 mimics. Furthermore, immunofluorescent staining of 2-day-old and 13-day-old hearts revealed that the cardiomyocytes at both developmental stages embodied the same level of IGF1 expression. Lastly, we discovered that miR-1 antagomir transfected into cardiomyocytes did not exert a positive proliferative effect on cardiomyocytes, which was inconsistent with our expectation. Despite this inconsistency, our study is the first to report miR-1’s ability to inhibit cardiomyocytes proliferation via the inhibition of CCND1 expression. Our findings also suggest that if we could identify a method to effectively inhibit miR-1 expression or stimulate CCND1 expression, then a novel therapeutic tool could be developed to reactivate cardiomyocyte proliferation after heart injury.
Conclusions
In this study, we have demonstrated that miR-1 was the most significantly upregulated miRNA in 13-day-old mouse heart compared with 2-day-old heart. We also revealed that miR-1 could repress cardiomyocyte G1/S phase transition, proliferation and viability. These results suggest that miR-1 plays an important role in inhibiting cardiomyocyte proliferation in the developing neonatal mouse heart by directly suppressing the cell-cycle regulator, CCND1.
Acknowledgments
Funding: This study was supported by General Research Fund (14100315). Guangdong Science and Technology Bureau International Science and Technology Collaboration Program (20130501c).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: All experimental protocol used in this study was approved by the Chinese University of Hong Kong Animal Care and User Committee and Department of Health (Ref No.: 13-2 in DH/HA&P/8/2/1 Pt.28). The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Huang Y, Harrison MR, Osorio A, et al. Igf Signaling is Required for Cardiomyocyte Proliferation during Zebrafish Heart Development and Regeneration. PLoS One 2013;8:e67266. [Crossref] [PubMed]
- Zhang R, Han P, Yang H, et al. In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature 2013;498:497-501. [Crossref] [PubMed]
- Tang MK, Kindler PM, Cai DQ, et al. Heart-type fatty acid binding proteins are upregulated during terminal differentiation of mouse cardiomyocytes, as revealed by proteomic analysis. Cell Tissue Res 2004;316:339-47. [Crossref] [PubMed]
- Liu Y, Tang MK, Cai DQ, et al. Cyclin I and p53 are differentially expressed during the terminal differentiation of the postnatal mouse heart. Proteomics 2007;7:23-32. [Crossref] [PubMed]
- Porrello ER, Mahmoud AI, Simpson E, et al. Transient Regenerative Potential of the Neonatal Mouse Heart. Science 2011;331:1078-80. [Crossref] [PubMed]
- Eulalio A, Mano M, Ferro MD, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 2012;492:376-81. [Crossref] [PubMed]
- Wahlquist C, Jeong D, Rojas-Munoz A, et al. Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature 2014;508:531-5. [Crossref] [PubMed]
- Feng Y, Yu X. Cardinal roles of miRNA in cardiac development and disease. Science China Life Sciences 2011;54:1113-20. [Crossref] [PubMed]
- Tang Y, Liu D, Zhang L, et al. Quantitative Analysis of miRNA Expression in Seven Human Foetal and Adult Organs. PLoS One 2011;6:e28730. [Crossref] [PubMed]
- Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet 2012;13:358-69. [Crossref] [PubMed]
- Feldman MY. Reactions of nucleic acids and nucleoproteins with formaldehyde. Prog Nucleic Acid Res Mol Biol 1973;13:1-49. [Crossref] [PubMed]
- Chen E, Tang MK, Yao Y, et al. Silencing BRE Expression in Human Umbilical Cord Perivascular (HUCPV) Progenitor Cells Accelerates Osteogenic and Chondrogenic Differentiation. PLoS One 2013;8:e67896. [Crossref] [PubMed]
- Gan J, Sonntag H-J. Integrative Analysis of the Developing Postnatal Mouse Heart Transcriptome. PLoS One 2015;10:e0133288. [Crossref] [PubMed]
- Di Stefano V, Giacca M, Capogrossi MC, et al. Knockdown of Cyclin-dependent Kinase Inhibitors Induces Cardiomyocyte Re-entry in the Cell Cycle. J Biol Chem 2011;286:8644-54. [Crossref] [PubMed]
- Cheng RK, Asai T, Tang H, et al. Cyclin A2 Induces Cardiac Regeneration After Myocardial Infarction and Prevents Heart Failure. Circ Res 2007;100:1741-8. [Crossref] [PubMed]
- Nakajima K, Inagawa M, Uchida C, et al. Coordinated regulation of differentiation and proliferation of embryonic cardiomyocytes by a jumonji (Jarid2)-cyclin D1 pathway. Development 2011;138:1771-82. [Crossref] [PubMed]
- Tane S, Kubota M, Okayama H, et al. Repression of Cyclin D1 Expression Is Necessary for the Maintenance of Cell Cycle Exit in Adult Mammalian Cardiomyocytes. J Biol Chem 2014;289:18033-44. [Crossref] [PubMed]
- Go AS, Mozaffarian D, Roger VL, et al. Heart Disease and Stroke Statistics—2014 Update: A Report From the American Heart Association. Circulation 2014;129:e28-e292. [PubMed]
- Lewis BP, Burge CB, Bartel DP. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell 2005;120:15-20. [Crossref] [PubMed]
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development 2005;132:4653-62. [Crossref] [PubMed]
- Cheng Y, Ji R, Yue J, et al. MicroRNAs Are Aberrantly Expressed in Hypertrophic Heart. Am J Pathol 2007;170:1831-40. [Crossref] [PubMed]
- Sluijter JP, van Mil A, van Vliet P, et al. MicroRNA-1 and -499 Regulate Differentiation and Proliferation in Human-Derived Cardiomyocyte Progenitor Cells. Arterioscler Thromb Vasc Biol 2010;30:859-68. [Crossref] [PubMed]
- Li Q, Song XW, Zou J, et al. Attenuation of microRNA-1 derepresses the cytoskeleton regulatory protein twinfilin-1 to provoke cardiac hypertrophy. J Cell Sci 2010;123:2444-52. [Crossref] [PubMed]
- Zhao Y, Ransom JF, Li A, et al. Dysregulation of Cardiogenesis, Cardiac Conduction, and Cell Cycle in Mice Lacking miRNA-1-2. Cell 2007;129:303-17. [Crossref] [PubMed]
- Liu L, Yuan Y, He X, et al. MicroRNA-1 upregulation promotes myocardiocyte proliferation and suppresses apoptosis during heart development. Mol Med Rep 2017;15:2837-42. [Crossref] [PubMed]
- Kwon C, Han Z, Olson EN, et al. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A 2005;102:18986-91. [Crossref] [PubMed]


