
Genetic variants and clinical significance of pediatric acute lymphoblastic leukemia
Introduction
Acute lymphoblastic leukemia (ALL), the most common childhood tumor, results in the malignant transformation of lymphoid progenitor cells, with more than 80% originating from B-cell progenitors (1). Childhood ALL develops more frequently in boys than in girls (male: female ratio, 55% to 45%) with the peak incidence occurring at 2 to 5 years of age (2). With intensified chemotherapy, remarkable progress has been made in the treatment, and the five-year overall survival (OS) rate can reach 85% to 90% in pediatric ALL (2,3). However, relapse occurs in approximately 20% of children and is associated with a high rate of treatment failure and death, particularly when occurring in the first 18 months of therapy. It remains the leading cause of cancer-related death in children and young adults (4-6).
Cytogenetic alterations and molecular abnormalities are frequent, and several molecular markers have been identified to stratify risk and predict prognosis, as they play key roles in ALL pathogenesis. A few genetic alterations have been shown to have clinical significance and different mutation distributions have been revealed; for example, rare germline mutations in the genes PAX5 (7) and ETV6 (8) were found to be linked to familial leukemia. PHF6 mutants had higher mutation prevalence in males (32.0% vs. 2.5%) in T-cell ALL (T-ALL) (9); Ras mutations (KRAS, NRAS, FLT3, PTPN11, NF1) are recurrent in pediatric B-cell ALL (B-ALL) and relapsed ALL patients, and their mutations may lead to prednisolone resistance (10,11); TP53 mutations mostly occur in low hypodiploid (chromosome <44) and are associated with relapse (12,13); SETD2 mutations often exist in relapsed ALL patients that are resistant to DNA-damaging chemotherapy agents (e.g., cytarabine, 6-TG, doxorubicin) and with a poor long-term survival (14-16); CREBBP mutations in the histone acetyltransferase (HAT) domain confer glucocorticoid resistance (13,17,18); NT5C2 mutations confer resistance to 6-mercaptopurine and 6-thioguanine (19,20); PRPS1 mutations are associated with thiopurine resistance (21).
However, the relevance of genetic alterations on disease phenotypes and clinical outcomes is largely unknown. Thus, understanding the genetic variants and clinical characteristics combined with evaluating the therapeutic effect and prognosis, may help us to explore the clinical significance and molecular pathogenesis. This may even improve the prognostic prediction for patients and help inform the selection of specific therapies. Also, with the advance of next-generation sequencing (NGS) technologies, simultaneous sequencing of multiple cancer-related genes through multiplex assay panels has become a more time and cost-efficient genetic testing strategy than single gene testing.
In this study, we intended to investigate the possible associations between genetic alterations and clinical phenotypes in Chinese pediatric patients with ALL, focusing on the influence of gene mutations on clinical significance and outcome.
Methods
Ethical compliance
Informed consent was obtained in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Children’s Hospital of Fudan University (No. [2015]005), Shanghai, China.
Patients and samples
We evaluated a total of 140 Chinese pediatric patients with ALL enrolled consecutively, who had been diagnosed and treated in the children’s hospital of Fudan University in China between January of 2015 and December of 2017. The diagnosis was based on the World Health Organization’s classification and patients were treated using the CCCG-ALL-2015 protocol, which was modified from St. Jude Children’s Research Hospital Total-XV protocol for newly diagnosed patients with ALL (Chinese protocol). Morphological, immunophenotyped and cytogenetic analyses were performed at the time of diagnosis. Bone marrow (BM) biopsy provided conclusive proof of ALL, typically with ≥20% of blast cells being leukemic lymphoblasts. Immunophenotypic determination of lineage commitment and developmental stage by flow cytometry are essential for correct diagnosis of ALL, while minimal residual disease (MRD) is also monitored by flow cytometry at day 19 and day 46. Cytogenetic analysis can be stratified according to ploidy, number of sets of chromosomes in the cell, and specific genetic abnormalities, such as translocations. The transcripts of BCR-ABL1, ETV6-RUNX1, TCF3-PBX1, and SIL-TAL1 fusion genes, along with MLL rearrangement (MLLr), were detected with a reverse transcriptase polymerase chain reaction (RT-PCR) or fluorescence in situ hybridization (FISH), as previously described (22).
Retrospective evaluation included an assessment of underlying disease, clinical manifestations, laboratory findings, treatment, and outcomes. Laboratory findings included peripheral blood examination, serum ferritin (SF), lactate dehydrogenase (LDH), BM morphology, flow cytometry, cerebrospinal fluid examination (CSF); imaging tests for testicular invasion and lymph nodes included B ultrasound examination and computerized tomography (CT).
BM samples were collected at the time of diagnosis and matched with remission samples or fingernails as germline controls. Genomic DNA was extracted from cell pellets using the DNAeasy Blood and Tissue Kit (Qiagen, USA). DNA was quantified using a Qubit Fluorometer (Life Technologies, USA), and DNA integrity was assessed by agarose gel electrophoresis.
Targeted capture sequencing and mutation analysis
Mutation analysis was performed by deep sequencing of 950 targeted exons genes related to cancer with sufficient reads coverage using a probe sequence capture array of Roche (http://www.nimblegen.com/products/seqcap/ez/v2/index.html) to enrich the exonic DNA (Joy Orient, China). The samples were sequenced on an Illumina Hiseq2500, and two parallel reactions were performed for each sample. After sequencing, BclToFastq (Illumina) was used to process the raw image files for base calling, and low-quality variations (quality score ≥20, Q20) were filtered. Cleaned reads were aligned to NCBI human reference genome (hg19) using Bowties2 (version 2.3.1), Samtools (version 1.1) and GATK (version 3.1.1) were used to analyze the single nucleotide variants (SNVs) and insertion or deletion in the sequence. Variants were queried against publicly available datasets such as 1,000 Genomes, NHLBI GO Exome Sequencing Project (ESP), and Exome Aggregation Consortium (ExAC) to filter out common polymorphisms [minor allele frequency (MAF) >0.01]. Synonymous changes and single nucleotide polymorphisms (SNPs) that MAF determined to be higher than 5% were removed (http://www.ncbi.nlm.nih.gov/projects/SNP). Nonsynonymous changes and small indels were filtered using SIFT software (version 1.03), Polyphen2 (version 2.2.2), PROVEAN (version 1.1.3), and MutationTaster 2. Variants associated with no-functional or truncating-proteins were classified as deleterious mutations. Deleterious mutations included stop-gain mutations, frameshift mutations, and splice site mutations. To identify candidate driver mutations, we filtered events, and the filtering criteria were a minimum coverage ≥10, minimum tumor variant frequency ≥0.10, normal variant frequency ≤0.05; any two prediction algorithms predicted to be deleterious or identified as recurrent in COSMIC were considered as candidate driver genes. Variants presented only in tumor samples were classified as somatic mutations.
In this study, we analyzed the association between clinical phenotypes and significant somatic mutations in 18 genes, these mutated genes occurred in more than 3 patients and were only limited to sequence analysis.
Statistical analysis
SPSS 24.0 (SPSS, Chicago, IL, USA) was used for statistical analysis. Comparisons of the categorical variables and continuous parameters were determined by Pearson’s Chi-square test or Fisher’s exact test. The Kaplan-Meier method was used to calculate the estimates of survival probability, which were compared by the log-rank test. Cox proportional hazards regression was used to analyze the possible factors of a recurrence by using a backward-selection stepwise modeling process. Relapse free survival (RFS) was defined as the time from a complete remission to relapse, or when follow-up was terminated. Two-sided P<0.05 was considered statistically significant.
Results
Comparisons of clinical characteristics between B-ALL and T-ALL
Among the 140 pediatric ALL patients enrolled in the analysis, 81.4% (n=114) were B-cell ALL (B-ALL) and 18.6% (n=26) were T-ALL. They included 86 males and 54 females, and the mean diagnosis age was 4.9 (range, 0.3–13.8 years). When compared with B-ALL patients, we found that newly diagnosed T-ALL patients had higher initial white blood cell (WBC) counts (34.8×109/L vs. 7.6×109/L, P=0.046), higher hemoglobin level (median 103 vs. 74.7 g/L, P=0.02), higher incidence of mediastinal mass (26.9% vs. 1.8%, P<0.001), higher LDH level (LDH ≥448 IU/mL, 86.4% vs. 49.6%, P=0.001) and easily occurring relapse (23.1% vs. 7.0%, P=0.036). There were no differences in gender, central nervous system (CNS) leukemia, testicular invasion at diagnosis, and treatment response of day 19 or day 46. Clinical characteristics of ALL patients at diagnosis are described in Table 1.
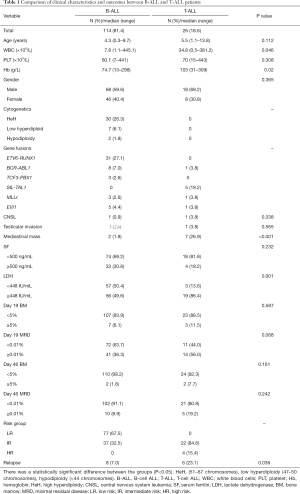
Full table
Recurrent deleterious mutations in pediatric ALL
Most of the ALL patients (n=138, 98.6%) harbored somatic mutations, and a majority, 72.9% (n=102) of ALL patients, carried more than one deleterious mutation. Even though T-ALL patients had higher mutational numbers in both coding mutations (average 6 vs. 8, P=0.267) and driver mutations (average 1 vs. 2, P=0.179), there was no significant difference between them (Figure 1A,B).
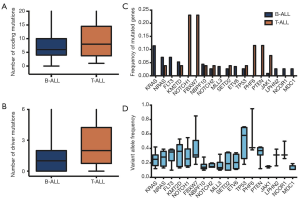
In all mutations, we found 18 deleterious mutations occurred in more than three ALL patients, and recurrently mutated genes with a mutation prevalence over 5% included KRAS (9.3%), NRAS (6.4%), FLT3 (5.7%), and KMT2D (5.0%) in childhood ALL. Genetic profiling was substantially different between B-ALL and T-ALL, including KRAS (11.4%), NRAS (7.0%), FLT3 (7.0%), and KMT2D (5.3%) which were frequently mutated in B-ALL. Meanwhile, NOTCH1 (23.1%), FBXW7 (23.1%), PHF6 (11.5%), and PTEN (11.5%) were enriched in T-ALL (Figure 1C). Among these mutations, JAK1 mutations showed a low allelic burden and were considered more frequently to be from a subclone than a clone, suggesting that these mutations were more likely to be late events than founder alterations (Figure 1D, Table S1). However, some mutations co-existed in the same patients; for example, NRAS/KRAS occurred in one B-ALL patient, while NOTCH1/FBXW7 and NOTCH1/ PHF6 existed in two T-ALL patients (Figure 2).
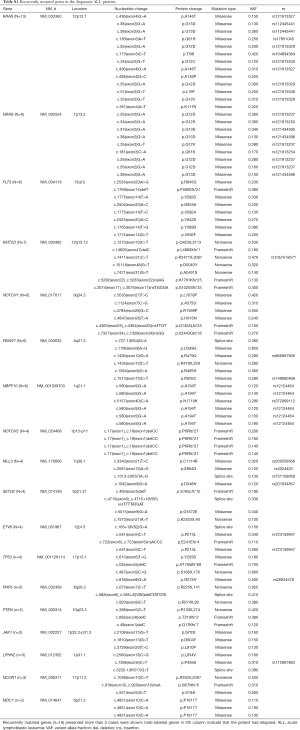
Full table
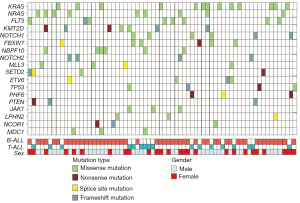
Associations of genetic features with clinical characteristics
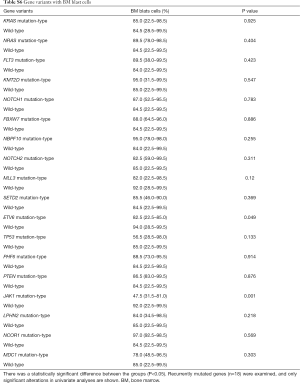
The association between genetic variations and clinical characteristics in 140 ALL patients were analyzed. In our study, there were no significant differences between clinical features and the number of somatic mutations. Furthermore, gene fusion-positive patients often co-existed with gene mutations (Table S2). Considering that clinical significance for deleterious mutations may exist, we evaluated the effects of individual alterations on the clinical features and treatment responses. Within the limits presented by the small number of subjects analyzed, we found a high rate were positive for KRAS mutations in our cohort, corresponding to an 11.4% incidence in B-ALL; however, no clinical correlation was found in this subgroup of patients. Interestingly, among other recurrently mutant genes, we found that SETD2 and TP53 mutations were more frequent in females (7.4%, P=0.041), and SETD2 mutants were older than SET2D wild patients (5.5 vs. 4.5 years, P=0.041). However, TP53 mutants were not characterized by an older median age as previously published by Stengel et al. (23). Meanwhile, NOTCH1 or SETD2 mutants were often found with higher initial WBC counts (≥50×109/L, P=0.047 and P=0.044 respectively) for newly diagnosed ALL patients, but ETV6 or JAK1 mutants had lower primary BM blast cells. It seems that the number of initial WBC in peripheral blood is not associated with the number of primitive blast cells in the BM.
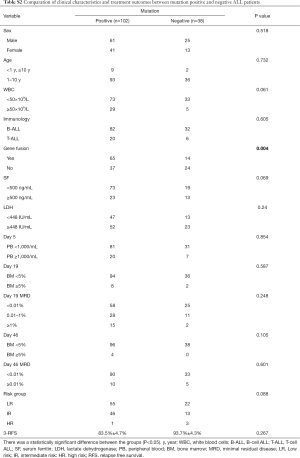
Full table
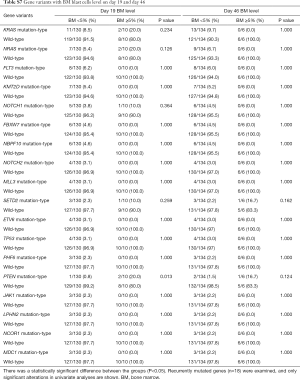
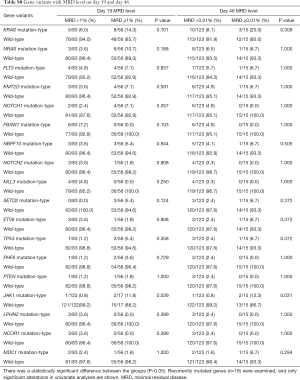
Furthermore, when comparing genetic alterations and treatment responses, it was revealed that PTEN mutants with higher BM blast cells at day 19 (20% vs. 0.8%, P=0.013) and JAK1 mutants had higher MRD level on both day 19 and day 46 (day 19 MRD ≥1%, P=0.039; day 46 MRD ≥0.01%, P=0.031) (Tables S3-S8). No other correlation with clinical features, such as gender, age, initial WBC counts, the percentage of blasts at diagnosis, and treatment outcomes, emerged from this analysis. The clinical significance of 18 mutated genes is summarized in Tables 2,S3-S8.
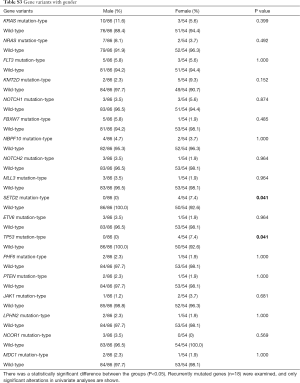
Full table
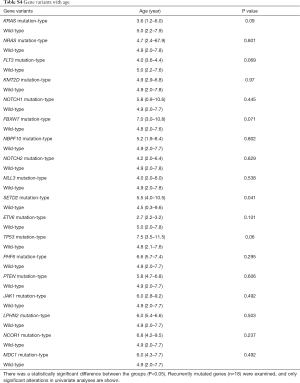
Full table
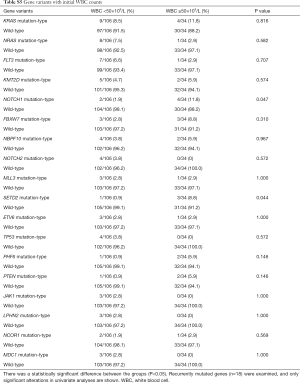
Full table
Full table
Full table
Full table
Full table
Associations of genetic features with clinical outcomes
At the deadline of December 31st, 2017, in all 14 patients (Table S9), a relapse occurred, including 4 cases of early relapse (within more than 18 months from the first remission but less than 6 months after chemotherapy finished) and 10 cases of very early relapse (within less than 18 months from the first remission). In the treatment program, it was divided into low-risk and medium-high-risk protocol. Therefore, we further analyzed the relationship between individual alterations, clinical characteristics and RFS. Indeed, the frequency of relapse in patients mutated for TP53 (50%) and NOTCH1 (33.3%) was higher than other mutations. We identified six NOTCH1 mutations, including 4 novel missenses mutations (L1678P, A375G, R1598P and I1616N) and 2 frameshift mutations (Q1455 Lfs*25, V2443Gfs*35) in primary diagnosed ALL patients, two pediatric patients with L1678P or A375G relapsed (Tables S1,S9) and with a shorter 3-year RFS rate 33.3% (P=0.006) (Figure 3A). Similarly, 4 patients carrying a TP53 alteration entered clinical remission after induction therapy, but 2 patients with H179Mfs*68 or R273H suffered an early relapse, and the 3-year RFS rate was 33.3% (Figure 3B).
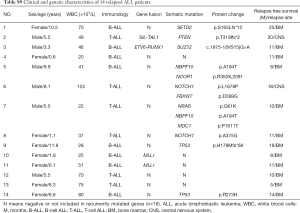
Full table

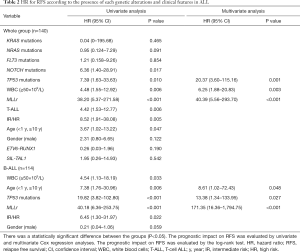
In a univariate analysis, somatic mutations involving NOTCH1 and TP53 were significantly associated with a poor outcome. In addition, several clinical prognostic factors were evaluated, and we found that WBC counts (≥50×109/L), MLLr, T-ALL, diagnosed age (<1 year or ≥10 years) as well as risk stratification (intermediate-high risk) were also significant predictors of an inferior survival (Table 2). In particular, MLLr was strongly predictive, with an odds ratio of 38.20 (5.37–271.58). Finally, we evaluated the relative effects of different mutations and clinical features together using Cox proportional hazards modeling with stepwise variable selection, incorporating initial WBC counts (≥50×109/L), MLLr, T-ALL, age, NOTCH1, TP53 mutations and intermediate-high risk as covariates. We found that higher initial WBC counts (≥50×109/L), TP53 mutations, and MLLr were independently associated with a shorter RFS, of which MLLr was the most significant predictor of clinical outcome of ALL patients with an odds ratio of 40.39 (5.56–293.70), suggesting a major role of these alterations in the progression of ALL (Table 2). Importantly, the effects of genetic alterations strongly depended on disease subtype: NOTCH1 mutations mainly occurred in T-ALL, and TP53 mutations were in B-ALL. Therefore, in the subsequent analyses, we stratified patients into B-ALL and T-ALL subtypes, incorporating subtype-specific clinical prognostic factors.
For B-ALL, TP53 mutations, together with age (<1 year or ≥10 years) and MLLr were independently associated with an adverse outcome, but high WBC counts (≥50×109/L) and intermediate-high risk were risk factor. Based on the number of these relevant risk factors they had, B-ALL patients were classified into three categories showing significantly different 1-year RFS rates (P<0.001): 100% for those with no risk factor, 91.3% for those with 1 risk factor, and 20% for those with ≥2 risk factors (Figure 3C). Thus, the evaluation of the molecular status of TP53 mutations, patient age, initial WBC counts, as well as MLLr would be informative in prognostication of B-ALL (Table 2).
However, because the number of T-ALL patients enrolled in this study was limited, and itself as an independent risk factor for recurrence in our cohort, no association was identified between T-ALL and genetic alterations.
Discussion
By analyzing clinical characteristics and genotyping data, we intended to demonstrate the clinical effects of genetic alterations, and looked to understand the significance of genetic profiling for prognostication in pediatric ALL.
The mutation profiling which occurred in our pediatric ALL was comparable to that reported in previous studies (3-6), whereas the incidence of NOTCH1 mutation was lower than the incidence reported in some studies from western populations and Chinese populations (5,24). Due to the detection of sequence mutations in ALL being insufficient, large deletion, amplification, rearrangement, and translocation should be warranted in the future.
Among the recurrent alterations, TP53 mutations and MLLr were a powerful predictor for an adverse outcome in B-ALL and pediatric ALL. The poor prognosis of MLLr is well-recognized, and the basis for risk stratification in chemotherapy regimens, small molecule inhibitor pinometostat, has entered phase 1 clinical trials in both adult and pediatric MLLr leukemia, with the expectation that it will improve the prognosis of patients with MLLr in the future (25). TP53 mutations mostly presented in the low hypodiploid subtype of ALL, approximately 50% of which were germline in nature, and were independently associated with a short survival (12,13,23,26). In our cohort, 4 pathogenic mutations all occurred in the TP53 DNA-binding domain, which was likely to result in the ablation of the p53-mediated DNA damage response, thus forming a general resistance to antileukemia agents (27). Children with TP53 variants were at a higher risk of second cancers, with a 5-year cumulative incidence of 25.1% and TP53 mutation had independent prognostic value (28). Also, we found that TP53 mutations were common in females (P=0.041), and other research showed that TP53 mutation incidence increased with age (23,26). ALL patients carrying TP53 mutations entered clinical remission after induction therapy, but most of them suffered a very early relapse (less than 18 months from the first remission) resulting in a shorter RFS. These data show that the presence of mutated TP53 itself did not produce a primary resistance to the induction chemotherapy, but rather lead to a greater susceptibility to relapse, as previously reported (29).
SETD2 mutations easily occurred in female patients, which were with old age and higher initial WBC counts, but it was not predicted to be a prognostic factor. However, SETD2 mutations had functional involvement in relapsed ALL patients, whose loss lead to resistance to DNA-damaging chemotherapy agents (cytarabine, 6-TG, doxorubicin), caused chemotherapy resistance, and increased the mutation rate at the site of diminished H3K36me3, with poor long-term survival (14-16). As reported, SETD2 alterations were frequently associated with MLLr (22%), ETV6-RUNX1 (13%), and T-cell lymphoma (30,31), which is an essential interactor in the initiation and maintenance of MLLr leukemia (32); however, in our research, just one case co-existed with ETV6-RUNX1, and only one case occurred in T-ALL; thus, more data is needed to confirm this result.
JAK1 mutations were associated with higher day 19 and day 46 MRD level, even in T-ALL patients, with dramatically higher MRD level on day 19 (more than 10%) and a sustained MRD level of more than 0.01% on day 46, but JAK1 mutations showed low allelic burden and were considered more frequently to be from subclones. JAK1 mutations and rearrangement-activated JAKs were seen in Ph-like B-ALL, and in the CRLF2 rearrangements, had a poor outcome (33). The JAK1/JAK2 inhibitor, ruxolitinib, was approved for myeloproliferative neoplasm (MPN) patients (34), and preclinical activity was recently reported in models of childhood T-ALL (35), with HSP90 inhibition PU-H71 (36) showing preclinical efficacy in JAK1/JAK2 models of ALL. It is promising that the usage of these targeted drugs can decrease MRD level, intensify chemotherapy effect, increase induction remission, and improve the clinical outcome of these patients with JAK mutation.
In our cohort, there were two cases with NOTCH1 mutation who suffered a very early relapse (less than 18 months from the first remission) and showed a significant influence in RFS of ALL (P=0.006). NOTCH1 mutation was predicted to be a risk factor for early relapse in ALL patients. However, NOTCH1 mutations just occurred in T-ALL patients and did not have a definite relationship with T-ALL outcome. Several types of research revealed that pediatric patients with mutated NOTCH1 tended to show improved OS and EFS compared to those with wild-type NOTCH1 (24,37,38); however, other studies showed a poorer survival or no impact on T-ALL outcome (39). Therefore, using NOTCH1 mutations as an early relapse risk factor is still controversial, and requires further validation in larger prospective studies or T-ALL specific subtype.
Conspicuously, a molecular profile of subtypes, such as WBC (≥50×109/L), age (<1 year, ≥10 years), TP53 mutations, and MLLr, significantly predicted poor prognosis in B-ALL. Combination of these risk factor would enable us to identify a subset of patients who would benefit from more intensive treatment, such as combined chemotherapy, targeted therapy, and allogeneic hematopoietic stem cell transplantation (HSCT). These findings suggest that somatic mutations (e.g., TP53) combined clinical features help predict treatment outcomes and could improve the prognosis in B-ALL patients.
Conclusions
There are some limitations to the present study. The number of patients enrolled in the present study was relatively low; the follow-up time was too short, which may contribute to the discrepancies in the findings between our study and previous research. Also, due to the limitations of our technology, only detecting sequence mutations in ALL was insufficient, and large intragenic deletion, amplification, and translocation is warranted in the future. Thus, the prevalence of deleterious mutations among these genes may be underestimated.
In conclusion, using NGS to complete molecular profiling could potentially improve the prediction of prognosis in ALL patients and better guide therapy options, such as early intervention with combined chemotherapy and allogeneic HST, immune therapy or targeted therapy.
Acknowledgments
We thank all patients and families who participated in this study. Project Ai You Foundation Supporting Children with Cancer Program.
Funding: The research was funded by the Research Programs of the Shanghai Science and Technology Commission Foundation (No. 14411950603), Shanghai Municipal Commission of Health and Family Planning (No. 201740011).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: Informed consent was obtained in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Children’s Hospital of Fudan University (No. [2015]005), Shanghai, China.
References
- Inaba H, Greaves M, Mullighan CG. Acute lymphoblastic leukemia. Lancet 2013;381:1943-55. [Crossref] [PubMed]
- Hunger SP, Mullighan CG. Acute Lymphoblastic Leukemia in Children. N Engl J Med 2015;373:1541-52. [Crossref] [PubMed]
- Ding LW, Sun QY, Tan KT, et al. Mutational landscape of pediatric Acute lymphoblastic leukemia. Cancer Res 2017;77:390-400. [Crossref] [PubMed]
- Mullighan CG. Genomic characterization of childhood acute lymphoblastic leukemia. Semin Hematol 2013;50:314-24. [Crossref] [PubMed]
- Tasian SK, Hunger SP. Genomic characterization of pediatric acute lymphoblastic leukemia:an opportunity for precision medicine therapeutics. Br J Haematol 2017;176:867-82. [Crossref] [PubMed]
- Roberts KG, Mullighan CG. Genomics in acute lymphoblastic leukaemia: insights and treatment implications. Nat Rev Clin Oncol 2015;12:344-57. [Crossref] [PubMed]
- Shah S, Schrader KA, Waanders E, et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet 2013;45:1226-31. [Crossref] [PubMed]
- Zhang MY, Churpek JE, Keel SB, et al. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat Genet 2015;47:180-5. [Crossref] [PubMed]
- Van Vlierberghe P, Palomero T, Khiabanian H, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet 2010;42:338-42. [Crossref] [PubMed]
- Ariës IM, van den Dungen RE, Koudijs MJ, et al. Towards personalized therapy in pediatric acute lymphoblastic leukemia: RAS mutations and prednisolone resistance. Haematologica 2015;100:e132-6. [Crossref] [PubMed]
- Irving J, Matheson E, Minto L, et al. Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood 2014;124:3420-30. [Crossref] [PubMed]
- Holmfeldt L, Wei L, Diazflores E, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet 2013;45:242-52. [Crossref] [PubMed]
- Ma X, Edmonson M, Yergeau D, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukemia. Nat Commun 2015;6:6604-16. [Crossref] [PubMed]
- Mar BG, Bullinger LB, McLean KM, et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in pediatric acute lymphoblastic leukaemia. Nat Commun 2014;5:3469-80. [Crossref] [PubMed]
- Wang Q, Cheng T. Evidences for mutations in the histone modifying gene SETD2 as critical drivers in leukemia development. Sci China Life Sci 2014;57:944-6. [Crossref] [PubMed]
- Mar BG, Chu SH, Kahn JD, et al. SETD2 alterations impair DNA damage recognition and lead to resistance to chemotherapy in leukemia. Blood 2017;130:2631-41. [Crossref] [PubMed]
- Mullighan CG, Zhang J, Kasper LH, et al. CREBBP mutations in relapsed acute lymphoblastic leukemia. Nature 2011;471:235-9. [Crossref] [PubMed]
- Malinowska-Ozdowy K, Frech C, Schönegger A, et al. KRAS and CREBBP mutations: a relapse-linked malicious liaison in childhood high hyperdiploid acute lymphoblastic leukemia. Leukemia 2015;29:1656-67. [Crossref] [PubMed]
- Tzoneva G, Perezgarcia A, Carpenter Z, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med 2013;19:368-71. [Crossref] [PubMed]
- Meyer JA, Wang J, Hogan LE, et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet 2013;45:290-4. [Crossref] [PubMed]
- Li B, Li H, Bai Y, et al. Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL. Nat Med 2015;21:563-71. [Crossref] [PubMed]
- Chen B, Wang YY, Shen Y, et al. Newly diagnosed acute lymphoblastic leukemia in China (I):abnormal genetic patterns in 1346 childhood and adult cases and their comparison with the reports from Western countries. Leukemia 2012;26:1608-16. [Crossref] [PubMed]
- Stengel A, Schnittger S, Weissmann S, et al. TP53 mutations occur in 15.7% of ALL and are associated with MYC-rearrangement, low hypodiploidy, and a poor prognosis. Blood 2014;124:251-8. [Crossref] [PubMed]
- Yuan L, Lu L, Yang Y, et al. Genetic mutational profiling analysis of T cell acute lymphoblastic leukemia reveal mutant FBXW7 as a prognostic indicator for inferior survival. Ann Hematol 2015;94:1817-28. [Crossref] [PubMed]
- Waters NJ. Preclinical Pharmacokinetics and Pharmacodynamics of Pinometostat (EPZ-5676), a First-in-Class, Small Molecule S-Adenosyl Methionine Competitive Inhibitor of DOT1L. Eur J Drug Metab Pharmacokinet 2017;42:891-901. [Crossref] [PubMed]
- Mühlbacher V, Zenger M, Schnittger S, et al. Acute lymphoblastic leukemia with low hypodiploid/near triploid karyotype is a specific clinical entity and exhibits a very high TP53 mutation frequency of 93%. Genes Chromosomes Cancer 2014;53:524-36. [Crossref] [PubMed]
- Irving JA, Enshaei A, Parker CA, et al. Integration of genetic and clinical risk factors improves prognostication in relapsed childhood B-cell precursor acute lymphoblastic leukemia. Blood 2016;128:911-22. [Crossref] [PubMed]
- Qian M, Cao X, Devidas M, et al. TP53 Germline Variations Influence the Predisposition and Prognosis of B-Cell Acute Lymphoblastic Leukemia in Children. J Clin Oncol 2018;36:591-9. [Crossref] [PubMed]
- Salmoiraghi S, Montalvo ML, Ubiali G, et al. Mutations of TP53 gene in adult acute lymphoblastic leukemia at diagnosis do not affect the achievement of hematologic response but correlate with early relapse and very poor survival. Haematologica 2016;101:e245-8. [Crossref] [PubMed]
- Zhu X, He F, Zeng H, et al. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat Genet 2014;46:287-93. [Crossref] [PubMed]
- McKinney M, Moffitt AB, Gaulard P, et al. The Genetic Basis of Hepatosplenic T-cell Lymphoma. Cancer Discov 2017;7:369-79. [Crossref] [PubMed]
- Skucha A, Ebner J, Schmöllerl J, et al. MLL-fusion-driven leukemia requires SETD2 to safeguard genomic integrity. Nat Commun 2018;9:1983-99. [Crossref] [PubMed]
- Harvey RC, Mullighan CG, Chen IM, et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood 2010;115:5312-21. [Crossref] [PubMed]
- Kleppe M, Kwak M, Koppikar P, et al. JAK-STAT pathway activation in malignant and nonmalignant cells contributes to MPN pathogenesis and therapeutic response. Cancer Discov 2015;5:316-31. [Crossref] [PubMed]
- Maude SL, Dolai S, Delgado-Martin C, et al. Efficacy of JAK/STAT pathway inhibition in murine xenograft models of early T-cell precursor (ETP) acute lymphoblastic leukemia. Blood 2015;125:1759-67. [Crossref] [PubMed]
- Kucine N, Marubayashi S, Bhagwat N, et al. Tumor-specific HSP90 inhibition as a therapeutic approach in JAK-mutant acute lymphoblastic leukemias. Blood 2015;126:2479-83. [Crossref] [PubMed]
- Gao C, Liu SG, Zhang RD, et al. NOTCH1 mutations are associated with favourable long-term prognosis in paediatric T-cell acute lymphoblastic leukaemia: a retrospective study of patients treated on BCH- 2003 and CCLG-2008 protocol in China. Br J Haematol 2014;166:221-8. [Crossref] [PubMed]
- Natarajan V, Bandapalli OR, Rajkumar T, et al. NOTCH1 and FBXW7 mutations favor better outcome in pediatric South Indian T-cell acute lymphoblastic leukemia. J Pediatr Hematol oncol 2015;37:e23-30. [Crossref] [PubMed]
- Zuurbier L, Homminga I, Calvert V, et al. NOTCH1 and/or FBXW7 mutations predict for initial good prednisone response but not for improved outcome in pediatric T-cell acute lymphoblastic leukemia patients treated on DCOG or COALL protocols. Leukemia 2010;24:2014-22. [Crossref] [PubMed]


