
Investigating the dysfunctional pathogenesis of Wilms’ tumor through a multidimensional integration strategy
Introduction
Epidemiological observations show that Wilms’ tumor (WT, also known as nephroblastoma) is the most common malignant kidney tumor in children (1). Moreover, 98% of cases occur in children who are under the age of 10 years and the disease is most common in children under the age of 3 years. The disease is mainly characterized by swelling of the abdomen, which may be accompanied by symptoms of digestive diseases such as abdominal pain, nausea, vomiting and loss of appetite. In addition to these core symptoms, there are problems such as divergent iris defects, urogenital abnormalities and mental retardation (2). Meanwhile, bone metastases and lung metastases occur in patients with severe prognosis and further infiltration of the inferior vena cava and right atrium could lead to sudden death (3,4). Clinicians, pathologists and radiation oncologists have taken corrective measures leading to a significant increase in the overall cure rate and long-term survival of WT patients (5). However, satisfactory results have not been achieved as poor prognosis, recurrent WT and treatment side effects are still high in some patients (6-8). Patients receiving chemotherapy exhibit some symptoms such as jaundice, ascites, hepatomegaly and pain in the right upper quadrant, causing hepatic vein occlusive disease (VOD) (9). Nevertheless, vincristine, doxorubicin (VDA), and whole lung radiotherapy (XRT) have some effects on both inflammatory myofibroblastoma (IMT) and fatal deficient blood enteritis. People with these types of diseases have low survival rate. Fortunately, researchers have focused on medical and biological aspects. Through unrelenting efforts, they have made great progress and brought out many valuable scientific results, including information on WT related genes and signal pathways, which have enhanced our understanding of the pathogenesis of this disease.
Studies have shown that WT is caused by genetic abnormalities, epigenetics, genetic mutations and environmental factors (10-12). Multiple studies have identified and confirmed that mutations in the Wilms tumor suppressor gene (WT1) are associated with the pathogenesis of WT. During embryonic development, WT1 induces differentiation of renal progenitor cells in response to Wnt signaling. In hereditary nephroblastoma, WT1 inhibits gene silencing of beta-catenin, leading to precancerous renal arrest (13). Furthermore, in the case of WT, the Wnt/beta-catenin pathway is highly activated which is influenced by AXIN2 (13-15). Additionally, Urbach et al. showed that Lin28/Let-7 pathway regulates kidney development and tumorigenesis (16). Bielen and other scientists have determined the role of PI3 and MAP kinases, and found that G1 cell cycle arrest and downstream signaling of cell death are potential drug targets. In addition, in WT, they also demonstrated the therapeutic potential of insulin-like growth factor (IGF). Based on the known kidney disease-related genes, a multi-factor regulation network was constructed through a systematic comprehensive strategy to determine the biological significance of WT dysfunction mechanism. By combining RNA-seq data, gene co-expression networks and differential expression analysis, dysfunctional co-expression modules were identified. In the modules, several genes were involved in various biological processes (BP) such as cell proliferation, differentiation, apoptosis and carcinogenesis. Moreover, the genes were also involved in WT-related signals such as Wnt/beta-catenin, mTOR/ERK and calcineurin. Consequently, based on transcriptional and post-transcriptional regulatory systems, we identified key regulators in each module. Statistical analysis showed that some regulators play an important role in the pathogenesis of WT by regulating multiple dysfunction modules. Finally, based on this multifactorial regulation network, potential drugs with potential pharmacological or toxicological effects on WT were predicted. This multi-factor co-expression network analysis not only helps to explore the relationship between gene modules and self-assembled components but also provides a valuable resource for further studies.
Methods
Data resources
RNA-seq datasets and miRNA-seq datasets of WT were downloaded from the Cancer Genome Atlas (TCGA) (17). RNA-seq datasets contained 120 primary WT patients and 6 normal individuals, while miRNA expression datasets to 126 primary WT patients and 6 normal individuals. This study was approved by the Clinical Research Ethics Committee of College of Medicine, Zhejiang University.
The NCBI Gene database contained 55 kidney cancer-related genes and 1,581 renal carcinoma-related genes (18). Additionally, there were 987 kidney cancer-related genes and 284 renal carcinoma-related genes in the OMIM database (19). After integration, a total of 2,301 disease-related genes were obtained.
STRING database (20) provides the most comprehensive view of the protein-protein interactions (PPIs) currently available and 10,514 proteins with 405,916 interactions (score >900) in human were collected from this database. Additionally, for transcriptional and post-transcriptional regulation, from TRVUST v2, 2,492 TFs (21) with 9,396 TF-target relationships in humans were collected. In RAID v2.0, 4,331 ncRNAs in humans were associated with 431,937 ncRNA-related interactions (value >0.5) (22). Moreover, information about drugs and drug-target were downloaded from the Drugbank database (23), which was used to predict potential drugs for WT.
Identification of functional modules
In order to explore the co-expression of 2,301 WT-related genes in 120 WT patients, a weighted gene co-expression network analysis (WGCNA) was performed on their gene expression data (24), and the functional modules in WT were determined. WGCNA is a systematic biological method used to describe genetic associations between different samples, and can be used to identify gene sets with high synergistic changes at expression levels. The algorithm uses the adjacency coefficient as a measure of the correlation between any gene pairs, i.e., the power exponent weight of the nth index of the correlation coefficient. The connections between genes in the network are subjected to scale-free networks, making the algorithm more biologically meaningful. This builds a hierarchical clustering tree based on correlation coefficients between genes, whose different branches represent different modules. Among them, the selection principle of soft threshold is to make the constructed network match with scale-free network characteristics. In the data analysis process, the soft threshold power =5, and screened the gene cluster with synergistic expression behavior as a module.
Differential expression analysis
Differential expression analysis on RNA-seq datasets and miRNA-seq datasets was performed using the R language limma package (25-27). First, the readings of RNA-seq and miRNA-seq were normalized using the voom functions. Then, the differentially expressed genes and miRNAs were identified using the lmFit and eBayes functions, respectively, at default parameters.
Determination of significant crosstalk module pairs
To interpret the relationships of modules involved in the pathogenesis of WT, we performed crosstalk analysis on modules using human PPI network as background set. Initially, the original PPI network was randomized 1,000 times while maintaining the size of each node in the network. Furthermore, the significance per module pair was calculated as:
n is the number of random network, in which the number of interactions between module pair is more than the number in the real network. If P≤0.05, the module pair was considered as significant crosstalk module pair.
Exploration of pivot regulators
At each module, the module’s pivot adjuster satisfied the following conditions: (I) the interaction between the governor and the module was greater than 2; (II) the interaction between the regulator and the hypergenometric test was less than 0.01. Similarly, based on drug-target information, potential drugs for WT were predicted.
Gene ontology (GO) and KEGG pathway enrichment analysis
In the enrichment analysis of genes, GO (P value cutoff =0.01, Q value cutoff =0.01) and KEGG pathway (P value cutoff =0.05, Q value cutoff =0.2) were performed with the R language Clusterprofiler package (21). In addition, the network function analysis were done by BinGO (22), a plug-in in Cytoscape.
Results
Identification of disease-related modules in WT
From the genetic perspective, functional modules represent a series of highly related genes. Meanwhile, genes in the same module may have similar biological functions and participate in the same biological pathway. At biological systems, finding a gene module with potential functions is actually a bridge to understand the differences between individual gene functions and global network characteristics. Therefore, identifying gene functional modules is important in understanding the molecular mechanisms of disease. To date, biologists have conducted extensive experiments and studies on kidney-related cancers, such as WT, and have identified related potential pathogenic genes. To gain insights into the underlying molecular mechanisms of WT, disease-related genes were first integrated from NCBI Gene database (18) and OMIM database (19), and a total of 2,301 genes were collected. Results showed that they had co-expression behaviors based on RNA-seq datasets from 120 patients with WT, and 23 functional modules were identified (Figure 1), possibly representing the underlying molecular mechanisms of kidney cancer-associated genes in WT pathogenesis. Moreover, differential expression analysis was performed on the RNA-seq data. The results showed that all of the module genes were differentially expressed (Figure 2), suggesting that these disease-related genes in modules might be involved in the pathogenesis of nephroblastoma. Therefore, these 23 modules were identified as disease-related dysfunctional modules.
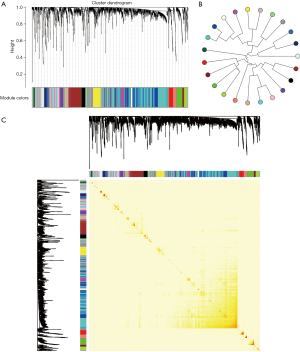
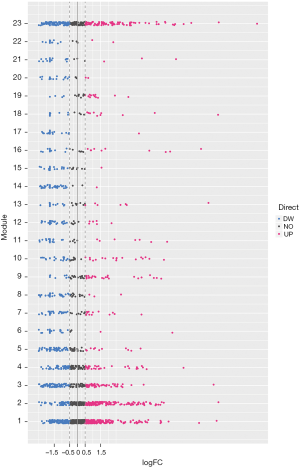
Functions and signaling pathways of involved modular gene
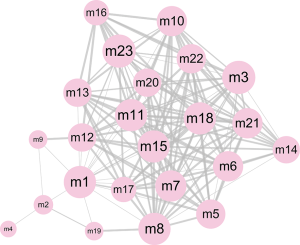
In the pathogenesis of genes involved in a disease, their function and pathway are important. Here, to further understand the biological characteristics of these modules, GO and KEGG pathway enrichment analysis on the 23 dysfunctional modules were performed. From a total of 580 cell composition (CC) entries, 355 molecular functional (MF) terms, 10,576 BP and 418 KEGG pathways (Figure S1), a number of BP were observed. These were mainly cell proliferation, migration, apoptosis, calcineurin, immune system regulation, regulation of the cell cycle and cancer signaling pathways, and various progenitor cells division. The KEGG pathway mainly includes Wnt/beta-catenin, JNK cascade and mTOR/ERK signaling pathways, which are involved in the development, progression and prognosis of WT. In addition, according to statistical analysis, up to 11 modules significantly participated in leukocyte differentiation, leukocyte migration, and protein kinase B signaling. This phenomenon indicates that WT is inextricably linked to the immune system, especially leukocyte immunity. On the other hand, according to the crosstalk relationship between modules, 146 pairs of modules with important crosstalk were identified (Figure 3). Through these crosstalk relationships, it can be considered that these disease-related genes not only have synergistic expression in the module, but also have mutual regulation between the modules, therefore forming a functional network to influence the process of WT.
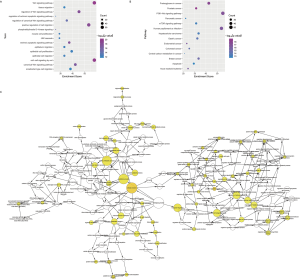
Pivot regulators and potential drugs correlated with disease-related modules
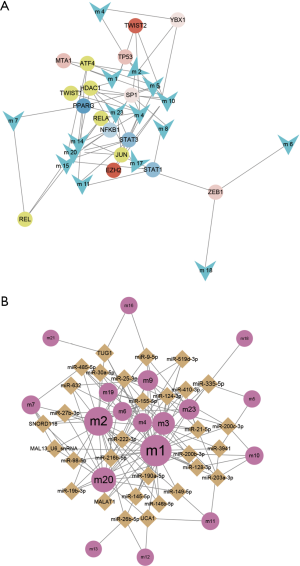
WT is a complex cancer, and the dysfunctional modules of WT are regulated by many factors. Based on the transcription and post-transcriptional regulation, a pivotal regulatory factor that significantly regulates dysfunction were identified. This included 157 transcription factors (TFs) with 2,569 TF-target relationships and 1,409 ncRNAs with 2,007 ncRNA-protein interactions (Figure 4). In addition, differentially expressed TFs and miRNAs in WT were identified based on RNA-seq and miRNA-seq datasets. Statistical analysis showed that the TF STAT3 significantly regulated six modules, and HDAC1, SP1, NFKB1 and RELA regulated five modules. Whereas miR-335-5p significantly regulates 7 modules, miR-21-5p, miR-25-3p, miR-9-5p, miR-27b-3p and TUG1 regulate 5 modules. By up-regulating miR-370, IL-6-activated STAT3 can inhibit the expression of WTX, which promotes the occurrence, proliferation and invasion of WT (24,28). SP1, a TF of WT1, mediates the incidence of WT (29). HDAC1 inhibits the expression of augWT1 and the corresponding target and this plays an important role in WT. Additionally, NFKB/Rel which up-regulates WT1 expression activates WT1 upstream regulatory cascade, promoting the development and progression of WT (30). MiR-21-5p was experimentally confirmed to be overexpressed in WT patients, and it promotes the migration and invasion of WT cells (31). Overall, these pivot regulators with high degree of certainty may have key effects on the regulation of the pathogenesis of WT.
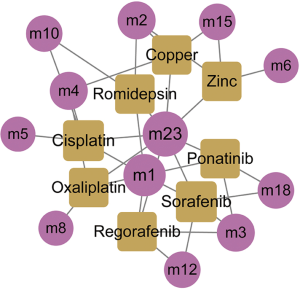
Based on the dysfunction mechanism of WT, it is desirable to explore potential therapeutic mechanisms for clinical and drug development, and to study effective treatments for reference. Here, based on the drug-target information from Drugbank (23), potential drugs for these dysfunctional modules were predicted. A total of 550 drugs with 721 drug-target interaction pairs were identified (Figure 5). These drugs have potential pharmacological or toxic effects on WT. Among them, cisplatin, sorafenib, copper and zinc have a significant regulation relationship with five modules, which may produce more significant effects or toxicity. Cisplatin has been used as a chemotherapeutic agent for WT. Sorafenib is an oral small molecule inhibitor, helpful in the treatment of rhabdomyosarcoma, nephroblastoma and hepatocellular carcinoma (HCC) (32,33). Studies have confirmed that ZFP346, ZNF224 and other zinc finger proteins act as transcriptional cofactors of WT1 in WT, leukemia and other cancers, and affect cancer progression (34,35). In addition, copper-bound monoamine oxidase (AOC1) is activated by WT1 and mediates polyamine breakdown, playing a key role in cell growth and proliferation of embryonic kidney and gonads (36). Exploring disease treatment mechanisms is the goal of most biomedical researchers. This study predicts that WT’s potential drugs lay a strong theoretical foundation for further exploration.
Discussion
Wilms tumor (WT) can be associated to the loss and mutation of the WT1 gene (located at 11p13) on chromosome 11. It may also be associated with proliferation and differentiation of mesenchymal cells in the posterior of kidney tissues (37,38). Although researchers have explored the etiology of WT at different aspects, the underlying pathogenesis is still unclear. In this study, 23 functional modules combined were identified using the WT patient’s RNA-seq data and 2,301 known kidney cancer-associated genes, and these genes were differentially expressed in WT. Enrichment analysis showed that these genes were involved in other validated pathways in addition to the common WT-related signaling pathways such as Wnt/β-catenin/Ras protein signal transduction, mTOR signaling pathway/positive regulation of ERK1 and ERK2 cascade (39-41). Studies have shown that in WT1, low expression, deletion and mutation could lead to increased expression of the IGF-1 receptor (IGF-IR), hence it is necessary to regulate the synthesis and secretion of IGF-II (42,43). Additionally, several studies have demonstrated that the phosphatidylinositol-3-kinase/Akt pathway is regulated by miR-19b, which mediates cell proliferation, cell cycle progression, migration and invasion, and participates in induction of glomerular proteinuria disease with activated macrophages in WT (44-46). Subsequently, 146 significant crosstalk relationships between modules were also observed, which bridged the gap between each potential dysfunctional mechanism. Moreover, by integrating TF targets and ncRNA-mRNA interactions, key regulators that significantly regulate dysfunction modules were identified. It highlights TFs STAT3, SP1, HDAC1, NFKB1, RELA, miR-335-5p, miR-21-5p, miR-25-3p, miR-9-5p, miR-27b-3p and lncRNA TUG1 and lncRNA TUG1 with higher levels. This study reveals the multidimensional effects of STAT3 in WT, and for instance, IL-6-activated STAT3 can inhibit expression of WTX. In addition, it is possible to up-regulate the expression of hypoxia-inducible factor-1alpha (HIF-1alpha) and vascular endothelial growth factor (VEGF). This promotes the development of WT, angiogenesis and the metastasis of cancer cells (24,28). SP1 has also been shown to be highly expressed in mesenchymal, early tubules, developing podocytes and mature glomeruli, and is a TF of WT1 which mediates WT (29,47). HDAC1 is recruited by the expressed WT1, which inhibits the expression of the corresponding target gene and plays an important role in WT (48). Moreover, a member of NFKB/Rel family up-regulates WT1 expression and activates WT1 upstream of the regulatory cascade, promoting the development and progression of WT (30). On the other hand, in various cancers including osteosarcoma, gastric cancer, non-small cell lung cancer, miR-335-5p was found to mediate tumor proliferation, migration and invasion. However, this has not been reported to play a role in WT (49-52). The predictive analysis of this study indicates its potential regulatory role in WT, but its basic molecular mechanism needs to be studied extensively. MiR-21-5p was experimentally confirmed to be overexpressed in WT patients, and the expression levels of phosphatidylinositol 3-kinase (PI3K) and phosphorylated protein kinase B (P-Akt) was regulated by PTEN targeting, promoting aggressive behavior such as migration and invasion of WT cells (31,53). In addition, miR-25-3p has also been reported to regulate the regulation of TGF-β1 expression, by inhibiting the adhesion of renal cancer cells, and promoting its proliferation and migration (54). miR-9-5p has been shown to promote the migration of mesenchymal stem cells (MSC) by activating the β-catenin signaling pathway, and affecting the formation and distribution of focal adhesions and the reorganization of F-actin, which may be potential pathogenesis of rhabdomyosarcoma nephroblastoma (55). LncRNA TUG1 has been reported to be overexpressed in cancers such as HCC, glioma, osteosarcoma and non-small cell lung cancer, and it likely promotes cell proliferation, growth, and apoptosis as well as enhance the permeability of blood tumor barrier (BTB). Thus, it could be identified as potential diagnostic biomarkers and therapeutic target (56-59). Although its role in WT has not been confirmed, based on the results of this study, TUG1 is highly likely to regulate certain key genes or pathways, which in turn mediates the development and progression of WT. Therefore, this can be used for further research studies.
Finally, based on these multifactorial regulated dysfunction modules and drug target information, potential drug predictions and statistical analyses were conducted. In 5 modules, cisplatin, sorafenib, zinc, and copper were found. However, cisplatin as a chemotherapeutic agent for WT, not only inhibits the growth of WT cells by inducing apoptosis, but also inhibits WT growth and invasion by promoting the binding of BAX protein to transcriptional activator 3 (STAT3) to target glucose regulatory protein 78 (GRP78). Sorafenib is an oral small molecule inhibitor that controls multiple kinases for tumor growth and angiogenesis. It can treat rhabdomyosarcoma, nephroblastoma and HCC that are unresponsive or intolerant to standard therapies. It has been established that Zinc can bind to certain proteins to form zinc finger proteins. Studies have confirmed that ZFP346, ZNF224 and other zinc finger proteins act as transcriptional cofactors of WT1 in WT, leukemia and other cancers, and they could have significant influence on cancer progression (34,35). Furthermore, it was observed that red blood cells from children with nephroblastoma have higher concentration of zinc and this is associated with zinc superoxide dismutase activity. On the other hand, AOC1 is activated by WT1 and mediates polyamine breakdown, playing a key role in cell growth and proliferation of embryonic kidney and gonads (36). Although the direct role of zinc and copper in WT has not been fully established, the results of this study suggest their potential roles.
In summary, this work reveals the multifactor-mediated dysfunctional network and characterizes the potential molecular mechanisms of WT. This not only clarifies the overall effect of related genes at a global level, but also improves the potential pathogenesis of WT, providing numerous candidate resources for future experimental verification. The prediction of potential drugs revealed the therapeutic mechanism of WT and as well provides candidate molecules for drug design and drug relocation studies of WT.
Acknowledgements
Funding: This study was supported by the National Natural Science Foundation of China (No. 81571953), the Major National S&T Projects for Infectious Diseases (No. 2018ZX10301401-005).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Al-Hussain T, Ali A, Akhtar M.. Wilms tumor: an update. Adv Anat Pathol 2014;21:166-73. [Crossref] [PubMed]
- Orawiec B, Młynarski W, Budzińska-Mikurenda M, et al. Sporadic aniridia and Wilm's tumor--a case report and review of recommendation for diagnostic approach in WAGR's syndrome. Klin Oczna 2010;112:321-3. [PubMed]
- Mohammadi A.. Recurrent pulmonary tumoral embolism and sudden death as the presenting symptom of Wilms' tumor. Tuberk Toraks 2011;59:271-5. [Crossref] [PubMed]
- Bond JV, Martin EC. Bone metastases in Wilms' tumour. Clin Radiol 1975;26:103-6. [Crossref] [PubMed]
- Spreafico F, Bellani FF. Wilms' tumor: past, present and (possibly) future. Expert Rev Anticancer Ther 2006;6:249-58. [Crossref] [PubMed]
- Sasso G, Greco N, Murino P, et al. Late toxicity in Wilms tumor patients treated with radiotherapy at 15 years of median follow-up. J Pediatr Hematol Oncol 2010;32:e264-7. [Crossref] [PubMed]
- Coppes MJ, Pritchard-Jones K. Principles of Wilms' tumor biology. Urol Clin North Am 2000;27:423-33. viii.. [Crossref] [PubMed]
- Suryanarayan K, Marina N.. Wilms' tumour. Optimal treatment strategies. Drugs 1998;56:598-605. [PubMed]
- Cesaro S, Spiller M, Sartori MT, et al. Veno-occlusive disease in pediatric patients affected by Wilms tumor. Pediatr Blood Cancer 2011;57:258-61. [Crossref] [PubMed]
- Deng C, Dai R, Li X, et al. Genetic variation frequencies in Wilms' tumor: A meta-analysis and systematic review. Cancer Sci 2016;107:690-9. [Crossref] [PubMed]
- Dumoucel S, Gauthier-Villars M, Stoppa-Lyonnet D, et al. Malformations, genetic abnormalities, and Wilms tumor. Pediatr Blood Cancer 2014;61:140-4. [Crossref] [PubMed]
- Scott RH, Murray A, Baskcomb L, et al. Stratification of Wilms tumor by genetic and epigenetic analysis. Oncotarget 2012;3:327-35. [Crossref] [PubMed]
- Tian F, Yourek G, Shi X, et al. The development of Wilms tumor: from WT1 and microRNA to animal models. Biochim Biophys Acta 2014;1846:180-7. [PubMed]
- Schweigert A, Fischer C, Mayr D, et al. Activation of the Wnt/beta-catenin pathway is common in wilms tumor, but rarely through beta-catenin mutation and APC promoter methylation. Pediatr Surg Int 2016;32:1141-6. [Crossref] [PubMed]
- Carraro DM, Ramalho RF, Maschietto M. Gene Expression in Wilms Tumor: Disturbance of the Wnt Signaling Pathway and MicroRNA Biogenesis. In: van den Heuvel-Eibrink MM. editor. Brisbane (AU): Wilms Tumor, 2016.
- Urbach A, Yermalovich A, Zhang J, et al. Lin28 sustains early renal progenitors and induces Wilms tumor. Genes Dev 2014;28:971-82. [Crossref] [PubMed]
- Tomczak K, Czerwinska P, Wiznerowicz M.. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. [Crossref] [PubMed]
- Brown GR, Hem V, Katz KS, et al. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res 2015;43:D36-42. [Crossref] [PubMed]
- Amberger JS, Bocchini CA, Schiettecatte F, et al. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res 2015;43:D789-98. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Maere S, Heymans K, Kuiper M.. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005;21:3448-9. [Crossref] [PubMed]
- Law V, Knox C, Djoumbou Y, et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res 2014;42:D1091-7. [Crossref] [PubMed]
- Cao X, Liu D, Yan X, et al. Stat3 inhibits WTX expression through up-regulation of microRNA-370 in Wilms tumor. FEBS Lett 2013;587:639-44. [Crossref] [PubMed]
- Law CW, Chen Y, Shi W, et al. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15:R29. [Crossref] [PubMed]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article3.
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Zhang LJ, Liu W, Gao YM, et al. The expression of IL-6 and STAT3 might predict progression and unfavorable prognosis in Wilms' tumor. Biochem Biophys Res Commun 2013;435:408-13. [Crossref] [PubMed]
- Cohen HT, Bossone SA, Zhu G, et al. Sp1 is a critical regulator of the Wilms' tumor-1 gene. J Biol Chem 1997;272:2901-13. [Crossref] [PubMed]
- Dehbi M, Hiscott J, Pelletier J.. Activation of the wt1 Wilms' tumor suppressor gene by NF-kappaB. Oncogene 1998;16:2033-9. [Crossref] [PubMed]
- Cui M, Liu W, Zhang L, et al. Over-Expression of miR-21 and Lower PTEN Levels in Wilms' Tumor with Aggressive Behavior. Tohoku J Exp Med 2017;242:43-52. [Crossref] [PubMed]
- Kim A, Widemann BC, Krailo M, et al. Phase 2 trial of sorafenib in children and young adults with refractory solid tumors: A report from the Children's Oncology Group. Pediatr Blood Cancer 2015;62:1562-6. [Crossref] [PubMed]
- Bregni M, Herr W, Blaise D, et al. Allogeneic stem cell transplantation for renal cell carcinoma. Expert Rev Anticancer Ther 2011;11:901-11. [Crossref] [PubMed]
- Montano G, Ullmark T, Jernmark-Nilsson H, et al. The hematopoietic tumor suppressor interferon regulatory factor 8 (IRF8) is upregulated by the antimetabolite cytarabine in leukemic cells involving the zinc finger protein ZNF224, acting as a cofactor of the Wilms' tumor gene 1 (WT1) protein. Leuk Res 2016;40:60-7. [Crossref] [PubMed]
- Schmitt J, Heisel S, Keller A, et al. Multicenter study identified molecular blood-born protein signatures for Wilms Tumor. Int J Cancer 2012;131:673-82. [Crossref] [PubMed]
- Kirschner KM, Braun JF, Jacobi CL, et al. Amine oxidase copper-containing 1 (AOC1) is a downstream target gene of the Wilms tumor protein, WT1, during kidney development. J Biol Chem 2014;289:24452-62. [Crossref] [PubMed]
- Pode-Shakked N, Dekel B.. Wilms tumor--a renal stem cell malignancy? Pediatr Nephrol 2011;26:1535-43. [Crossref] [PubMed]
- Gerald WL. The molecular genetics of Wilms tumor: a paradigm of heterogeneity in tumor development. Cancer Invest 1994;12:350-9. [Crossref] [PubMed]
- Yi Y, Polosukhina D, Love HD, et al. A Murine Model of K-RAS and beta-Catenin Induced Renal Tumors Expresses High Levels of E2F1 and Resembles Human Wilms Tumor. J Urol 2015;194:1762-70. [Crossref] [PubMed]
- Subbiah V, Brown RE, Jiang Y, et al. Morphoproteomic profiling of the mammalian target of rapamycin (mTOR) signaling pathway in desmoplastic small round cell tumor (EWS/WT1), Ewing's sarcoma (EWS/FLI1) and Wilms' tumor(WT1). PLoS One 2013;8:e68985. [Crossref] [PubMed]
- Hu Q, Gao F, Tian W, et al. Wt1 ablation and Igf2 upregulation in mice result in Wilms tumors with elevated ERK1/2 phosphorylation. J Clin Invest 2011;121:174-83. [Crossref] [PubMed]
- Qing RQ, Schmitt S, Ruelicke T, et al. Autocrine regulation of growth by insulin-like growth factor (IGF)-II mediated by type I IGF-receptor in Wilms tumor cells. Pediatr Res 1996;39:160-5. [Crossref] [PubMed]
- Werner H, Re GG, Drummond IA, et al. Increased expression of the insulin-like growth factor I receptor gene, IGF1R, in Wilms tumor is correlated with modulation of IGF1R promoter activity by the WT1 Wilms tumor gene product. Proc Natl Acad Sci U S A 1993;90:5828-32. [Crossref] [PubMed]
- Liu GL, Yang HJ, Liu B, et al. Effects of MicroRNA-19b on the Proliferation, Apoptosis, and Migration of Wilms' Tumor Cells Via the PTEN/PI3K/AKT Signaling Pathway. J Cell Biochem 2017;118:3424-34. [Crossref] [PubMed]
- Zhang M, Xue E, Shao W.. Andrographolide promotes vincristine-induced SK-NEP-1 tumor cell death via PI3K-AKT-p53 signaling pathway. Drug Des Devel Ther 2016;10:3143-52. [Crossref] [PubMed]
- Takano Y, Yamauchi K, Hayakawa K, et al. Transcriptional suppression of nephrin in podocytes by macrophages: roles of inflammatory cytokines and involvement of the PI3K/Akt pathway. FEBS Lett 2007;581:421-6. [Crossref] [PubMed]
- Discenza MT, Dehbi M, Pelletier J. Overlapping DNA recognition motifs between Sp1 and a novel trans-acting factor within the wt1 tumour suppressor gene promoter. Nucleic Acids Res 1997;25:4314-22. [Crossref] [PubMed]
- Lee KY, Jeon YJ, Kim HG, et al. The CUG-translated WT1, not AUG-WT1, is an oncogene. Carcinogenesis 2017;38:1228-40. [Crossref] [PubMed]
- Tang H, Zhu J, Du W, et al. CPNE1 is a target of miR-335-5p and plays an important role in the pathogenesis of non-small cell lung cancer. J Exp Clin Cancer Res 2018;37:131. [Crossref] [PubMed]
- Wang Y, Zeng X, Wang N, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer 2018;17:89. [Crossref] [PubMed]
- Zhang LL, Zhang LF, Guo XH, et al. Downregulation of miR-335-5p by Long Noncoding RNA ZEB1-AS1 in Gastric Cancer Promotes Tumor Proliferation and Invasion. DNA Cell Biol 2018;37:46-52. [Crossref] [PubMed]
- Wang Y, Yang T, Zhang Z, et al. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci 2017;108:859-67. [Crossref] [PubMed]
- Cui M, Liu W, Zhang L, et al. Clinicopathological parameters and prognostic relevance of miR-21 and PTEN expression in Wilms' tumor. J Pediatr Surg 2017;52:1348-54. [Crossref] [PubMed]
- Bogusławska J, Rodzik K, Poplawski P, et al. TGF-beta1 targets a microRNA network that regulates cellular adhesion and migration in renal cancer. Cancer Lett 2018;412:155-69. [Crossref] [PubMed]
- Li X, He L, Yue Q, et al. MiR-9-5p promotes MSC migration by activating beta-catenin signaling pathway. Am J Physiol Cell Physiol 2017;313:C80-93. [Crossref] [PubMed]
- Yun-Bo F, Xiao-Po L, Xiao-Li L, et al. LncRNA TUG1 is upregulated and promotes cell proliferation in osteosarcoma. Open Med (Wars) 2016;11:163-7. [Crossref] [PubMed]
- Huang MD, Chen WM, Qi FZ, et al. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol Cancer 2015;14:165. [Crossref] [PubMed]
- Cai H, Xue Y, Wang P, et al. The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget 2015;6:19759-79. [Crossref] [PubMed]
- Zhang EB, Yin DD, Sun M, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis 2014;5:e1243. [Crossref] [PubMed]


