
Genetics in systemic lupus erythematosus: entering the borough of cardiovascular risk
“You take delight not in a city’s seven or seventy-seven wonders, but in the answer that it gives to a question of yours.”—Italo Calvino, Invisible Cities, 1972.
Systemic lupus erythematosus (SLE) is a multi-organ autoimmune disease characterised by protean clinical manifestations due a constellation of pathogenic events with variable weight among different patients and in the same patient among different stages of the disease. Cardiovascular disease (CVD) constitutes a major cause of morbidity and mortality in patients with SLE and its prevalence is higher in SLE compared to the general population and even to other inflammatory diseases (1,2). Multiple pathogenic mechanisms are thought to underlie the development of vessel injury and ischaemia and include antiphospholipid antibodies (aPL), acquired dysfunction of the coagulation cascade, accelerated atherosclerosis and aberrant interactions among the vessel walls and circulating immune players such as platelets and leukocytes (3,4). Nonetheless, the question what are the inflammation-specific drivers prompting the divergence of SLE ischaemic risk trajectories from those estimated by conventional cardiovascular risk factors remains largely unanswered (5,6).
During the last decades, genetic studies had a major role in the identification of crucial mechanisms accounting for innate and adaptive immune derangement in SLE. In particular, genome-wide association studies (GWAS) revealed a strong association of multiple polymorphisms in key genes for interferon-α (IFNα) antiviral-like responses, antigen recognition and presentation, co-stimulation as well as B- and T-cell activation with the development of SLE (7,8). Nonetheless, the hereditary factors involved in the development of specific SLE manifestations have only recently begun to be defined and explored and stand as borderless boroughs of a larger city within the perimeter of general SLE susceptibility (9-11). Research in this field promises to define the mechanistic bases for prognostic profiling of patients with SLE and for individualised treatment choices. In particular, the identification of possible genetic factors accounting for the disproportionate inflammation-related cardiovascular risk in SLE might have a major impact on patients’ quality of life and long-term survival (6).
A large multicentre study has recently ventured into this complex setting by assessing possible associations among a large array of single nucleotide polymorphisms (SNPs) and cardiovascular outcomes such as myocardial infarction (MI)/angina and transient ischaemic attack (TIA)/stroke (12). Leonard et al. identified three sets of SNPs segregating with ischaemic events and found functional correlates of potential cardiovascular relevance for two of them. In particular, three SNPs in the interleukin 19 (IL19) gene correlated with a composite MI/stroke outcome, whereas three SNPs in the signal recognition particle 54 - antisense 1 (SRP54-AS1) locus were associated with stroke/TIA. Interestingly, when patients with SLE were compared to the general population: (I) none of these SNPs associated with SLE as a general category, (II) no associations were found between these SNPs and CVD in the general population (12). These data suggest that the potential contribution of specific genetic variants to CVD in SLE and other autoimmune diseases (13-17) might be related to unique inflammatory events that are not part of the stereotyped cascade of alterations to vessel integrity and haemostasis characterising atherosclerosis and thrombophilia in the general population. Furthermore, these events seem not to coincide with mechanisms sustaining systemic inflammation. Consistently, the authors also showed that, at least for IL19 SNPs, an association with CVD was also observable in patients with rheumatoid arthritis, another immune-mediated disease characterised by increased ischaemic risk (12). Although SLE exploratory and validation cohorts were quite non-homogeneous and data regarding ischaemic outcomes were not available from a non-negligible fraction of patients, analysis of a large number of patients and controls constitute one of the major strength of the work by Leonard et al. By combining epidemiological data with functional evidence from gene expression profile, SLE biomarkers and predicted transcriptome from bioinformatics, the authors also constructed a model in which both IL19 and SRP54-AS1 risk alleles would affect IFNα-responses promoting aPL development, generalised lymphocyte and macrophage activation and possibly vessel injury (Figure 1). IFNα has a major role in the pathogenesis of SLE and has been associated to endothelial dysfunction and release of proinflammatory microparticles (18). Defective binding of IFNα-dependent transcription factors such as signal transducer and activator of transcription 1 (STAT1) to SRP54-AS1 and of Ikaros family zinc finger 3 (IKZF3, also known as Aiolos) to IL19 sequence has been postulated to affect the expression of neighbouring genes with potential downstream effects on local vascular and systemic inflammation. SRP54-AS1 affects the expression of FAM177A1 gene at the level of the arterial vessels and might contribute to vascular inflammation. However, the exact function of FAM177A1 gene is unknown. In a similar way, alterations of IL19 sequence and its interactions with transcription factors are thought to facilitate IL10 release, which, in turn, would have an enhancing effect on B-cell-mediated responses including autoantibody production. Consistently, patients bearing the IL19 risk allele had higher levels of plasma IL10 and aPL (12). In addition, the regulatory role of interferon regulatory factor 8 (IRF8), another potential determinant of CVD in SLE on genetic bases (13), towards IKZF3, further supports the relevance of the IFNα→IL19 gene→IL10 axis in conferring increased cardiovascular risk to patients with SLE (Figure 1). In summary, thanks to the combination of gene association analysis and functional evidence, Leonard et al. not only added another item to the map of genetic susceptibility to SLE and its subphenotypes, but also consistently contributed to address the question what are the specific inflammatory mechanisms linking SLE to CVD.
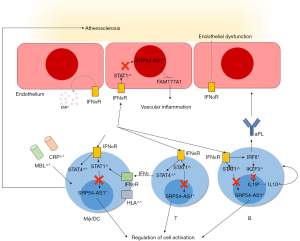
The two identified signalling pathway might thus become interesting future targets for therapeutic intervention. Notably, consistent with the evidence that only 10–15% of SLE-associated loci are located within protein-coding regions (8), IL19 and SRP54-AS1 SNPs did not affect protein translation, but only modulated the expression of other genes. Consequently, novel treatment approaches aiming at fine-modulating the complex interactions between key inflammation-related genes and their transcription regulators are warranted (19).
Acknowledgements
None.
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Fernandez-Gutierrez B, Perrotti PP, Gisbert JP, et al. Cardiovascular disease in immune-mediated inflammatory diseases: A cross-sectional analysis of 6 cohorts. Medicine (Baltimore) 2017;96:e7308. [Crossref] [PubMed]
- Lee JJ, Pope JE. A meta-analysis of the risk of venous thromboembolism in inflammatory rheumatic diseases. Arthritis Res Ther 2014;16:435. [Crossref] [PubMed]
- Liu Y, Kaplan MJ. Cardiovascular disease in systemic lupus erythematosus: an update. Curr Opin Rheumatol 2018;30:441-8. [PubMed]
- Ramirez GA, Efthymiou M, Isenberg DA, et al. Under crossfire: thromboembolic risk in systemic lupus erythematosus. Rheumatology (Oxford) 2018. [Epub ahead of print].
- Esdaile JM, Abrahamowicz M, Grodzicky T, et al. Traditional Framingham risk factors fail to fully account for accelerated atherosclerosis in systemic lupus erythematosus. Arthritis Rheum 2001;44:2331-7. [Crossref] [PubMed]
- Bakshi J, Segura BT, Wincup C, et al. Unmet Needs in the Pathogenesis and Treatment of Systemic Lupus Erythematosus. Clin Rev Allergy Immunol 2018;55:352-67. [Crossref] [PubMed]
- Bentham J, Morris DL, Graham DSC, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet 2015;47:1457-64. [Crossref] [PubMed]
- Chen L, Morris DL, Vyse TJ. Genetic advances in systemic lupus erythematosus: an update. Curr Opin Rheumatol 2017;29:423-33. [Crossref] [PubMed]
- Baqai T, Isenberg DA, Ioannou Y. Pathogenesis of neurocognitive and neuropsychiatric manifestations in childhood-onset lupus: an overview. Rheumatology Reports 2014;5:6. [Crossref]
- Govoni M, Bortoluzzi A, Padovan M, et al. The diagnosis and clinical management of the neuropsychiatric manifestations of lupus. J Autoimmun 2016;74:41-72. [Crossref] [PubMed]
- Mohan C, Putterman C. Genetics and pathogenesis of systemic lupus erythematosus and lupus nephritis. Nat Rev Nephrol 2015;11:329-41. [Crossref] [PubMed]
- Leonard D, Svenungsson E, Dahlqvist J, et al. Novel gene variants associated with cardiovascular disease in systemic lupus erythematosus and rheumatoid arthritis. Ann Rheum Dis 2018;77:1063-9. [Crossref] [PubMed]
- Leonard D, Svenungsson E, Sandling JK, et al. Coronary heart disease in systemic lupus erythematosus is associated with interferon regulatory factor-8 gene variants. Circ Cardiovasc Genet 2013;6:255-63. [Crossref] [PubMed]
- Cannarile F, Valentini V, Mirabelli G, et al. Cardiovascular disease in systemic sclerosis. Ann Transl Med 2015;3:8. [PubMed]
- Ohlenschlaeger T, Garred P, Madsen HO, et al. Mannose-binding lectin variant alleles and the risk of arterial thrombosis in systemic lupus erythematosus. N Engl J Med 2004;351:260-7. [Crossref] [PubMed]
- Svenungsson E, Gustafsson J, Leonard D, et al. A STAT4 risk allele is associated with ischaemic cerebrovascular events and anti-phospholipid antibodies in systemic lupus erythematosus. Ann Rheum Dis 2010;69:834-40. [Crossref] [PubMed]
- Lundstrom E, Gustafsson JT, Jonsen A, et al. HLA-DRB1*04/*13 alleles are associated with vascular disease and antiphospholipid antibodies in systemic lupus erythematosus. Ann Rheum Dis 2013;72:1018-25. [Crossref] [PubMed]
- Tyden H, Lood C, Gullstrand B, et al. Endothelial dysfunction is associated with activation of the type I interferon system and platelets in patients with systemic lupus erythematosus. RMD Open 2017;3:e000508. [Crossref] [PubMed]
- Trilling M, Bellora N, Rutkowski AJ, et al. Deciphering the modulation of gene expression by type I and II interferons combining 4sU-tagging, translational arrest and in silico promoter analysis. Nucleic Acids Res 2013;41:8107-25. [Crossref] [PubMed]


