Variations in mRNA and protein levels of Ikaros family members in pediatric T cell acute lymphoblastic leukemia
Introduction
Pediatric T cell acute lymphoblastic leukemia (T-ALL) is a heterogeneous disease in which the leukemic cells often resemble normal thymocyte populations. The phenotypic similarities between leukemic cells and normal thymocytes have led to the conclusion that patients might be classified according to the developmental stage that is most similar to the leukemic cells (1,2). For example, cells from some T-ALL cases lack TCRβ expression and are thought to be derived from early T cell precursors. During normal human T cell development, early T cell precursors are CD4−CD8− double negative (DN) thymocytes that express CD34. DN thymocytes can be divided into DN1, DN2, or DN3 cells, depending on their expression of CD38 and CD1a (3,4). During the DN stages, the cells commit to either the αβ or γδ T cell lineage (5,6). Cells from some T-ALL patients resemble uncommitted early T cell progenitors and likely emerge from the early DN stages. Cells from other patients express TCRγδ and resemble stages of γδ T cell development.
In other T-ALL cases, the leukemic cells express cytoplasmic TCRβ, but not surface CD3. During normal T cell development, cytoplasmic TCRβ+ cells are either immature single positive (ISP) CD4+ thymocytes or a subpopulation of CD4+CD8+ double positive (DP) thymocytes (7,8). ISP CD4+ thymocytes are a transition between the DN and DP stages (9). During the DP stage, thymocytes express TCRα, undergo positive or negative selection, and differentiate into mature single positive cells that can egress from the thymus and function as mature naive T cells.
There are conflicting data regarding the prognostic value of classifying pediatric T-ALL patients according to the phenotype of the leukemic cells. For example, some studies reported a poor prognosis for patients whose cells express either CD34 or surface TCR (10,11), while other studies found no differences in survival between groups (12,13). In this study, we used multi-parameter flow cytometry to demonstrate the degree of heterogeneity among T-ALL patients, perhaps explaining the challenges faced when attempting to correlate the phenotype of the leukemic cells with clinical outcomes.
Another parameter that might be used to categorize patients with T-ALL is the expression of Ikaros family members. The Ikaros family has been linked to T-ALL in mice (14-18) and humans (19-27) and consists of five highly homologous transcription factors: Ikaros, Helios, Aiolos, Eos, and Pegasus. Each family member has two zinc finger domains. The four N-terminal zinc fingers mediate DNA binding, and the two C-terminal zinc fingers are required for dimerization. Transcriptional activity requires dimerization and each family member is capable of dimerizing with each other family member (28-32).
Adding complexity to the study of Ikaros is that some family members undergo alternative splicing that results in the deletion of one or more DNA binding zinc fingers. Loss of one or two zinc fingers can result in more promiscuous DNA binding, but deletion of three or more zinc fingers can create dominant negative isoforms that block the ability of other family members to bind DNA (28,30-34). Some reports indicate that alternative splicing is common in T-ALL while other reports find it to be rare (19-27,35).
We compared mRNA and protein levels of each Ikaros family member in normal human thymocytes and cells from pediatric T-ALL patients. We identified patterns of Ikaros family mRNA levels that transcend surface phenotypes, suggesting that the key elements in investigating mechanisms of leukemogenesis lie in the function of Ikaros family proteins or the signaling pathways that regulate Ikaros family transcription.
Methods
Human tissue samples
Human thymus samples were obtained from children (0–18 years) that underwent corrective surgery at Children’s Mercy Hospital (Kansas City, MO, USA) for congenital cardiac defects after obtaining parent or guardian consent. Tissue samples were obtained in compliance with the Institutional Review Boards at Children’s Mercy Hospital and the University of Kansas Medical Center.
Twenty-four blood or bone marrow samples were obtained from newly diagnosed pediatric T-ALL patients prior to the initiation of anti-cancer therapy. Samples from patients diagnosed at Children’s Mercy Hospital were collected after obtaining parent or guardian consent. Leukemic cells were enriched by labeling the cells with anti-CD7-PE antibody and positively selecting with magnetic beads conjugated to anti-PE antibody (BD Biosciences, San Jose, CA, USA). Other samples were obtained as frozen samples through the Children’s Oncology Group (NCI Protocol AALL12B11). Each sample was divided into aliquots for flow cytometric, mRNA, and protein analysis.
Phenotypic analysis of T-ALL samples
The anti-human antibodies anti-CD1a-PerCP-Cy5.5, anti-CD1a-PECy5, anti-CD3-APCCy7, anti-CD3-APC-Alexa750, anti-CD4-Pacific Blue, anti-CD5-AF647, anti-CD7-PE, anti-CD8-BV785, anti-CD8-FITC, anti-CD27-HV500, anti-CD34-BV605, anti-CD34-PE, anti-CD34-PECy7, and anti-CD38-AF700, anti-CD44-PECy7, anti-CD45RO-PECy5, anti-CD69-BV650, anti-TCRγδ-FITC were purchased from Biolegend (San Diego, CA, USA). Samples were labeled with anti-CD1a, anti-CD3, anti-CD4, anti-CD5, anti-CD7, anti-CD8, anti-CD34, anti-CD38, and anti-TCRγδ antibodies. Most samples were also labeled with anti-CD27, anti-CD44, anti-CD45RO, and anti-CD69. Data were acquired using a BD LSR II (BD Biosciences) and analyzed using BD FACSDiva software (BD Biosciences).
FACS-purification of human thymocytes
DN2, DN3, and ISP thymocytes were obtained by depleting total thymocytes with magnetic beads conjugated to anti-CD8 and anti-CD3 (BD Biosciences). Remaining cells were divided into the DN2 (CD3−CD4−CD8−CD34+CD38+CD1a−), DN3 (CD3−CD4−CD8−CD34+CD38+CD1a+), and ISP (CD3−CD4+CD8−) populations using a FACSAria IIIu (BD Biosciences). For total DN or DP thymocytes, cells were labeled with anti-CD4 and anti-CD8 and CD4−CD8− or CD4+CD8+ thymocytes were FACS-purified.
Quantitative RT-PCR (qRT-PCR)
Total mRNA was isolated using the RNeasy Mini Kit or RNeasy Micro Kit (Qiagen, Valenica, CA, USA), converted to cDNA using the TaqMan® High Capacity RNA-to-cDNA™ kit (Applied Biosystems, Foster City, CA, USA), and amplified using TaqMan® Gene Expression Assays: Ikaros, Hs00958473_m1; Helios, Hs00212361_m1; Aiolos, Hs00232635_m1; Eos, Hs00223842_m1; Pegasus, Hs00223846_m1; and GAPDH, Hs03929097_g1 (Applied Biosystems). qRT-PCR was performed using a 7500 Fast Real-Time PCR System (Applied Biosystems). Each sample was run in triplicate. For normal thymocytes, at least five biological replicates were performed for each subset and each replicate was performed in triplicate. mRNA levels of each Ikaros family member were calculated relative to GAPDH and statistical significance was determined using the Log2(fold change).
Western blot analysis
Cell lysates prepared from 4×105–5×105 cells were separated by SDS-PAGE, transferred to nitrocellulose, and probed with antibodies against Ikaros, Aiolos, or p38 MAPK (all purchased from Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Bands were visualized using horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Inc.) and Pierce™ ECL Western Blotting Substrate (Life Technologies, Grand Island, NY, USA), and detected using an ImageQuant LAS-4000 gel imager (GE Healthcare systems, Pittsburgh, PA, USA).
Nested PCR
Total mRNA was reverse transcribed with AMV RT (Promega, Madison, WI, USA) before amplification with Taq DNA Polymerase (Fisher Scientific, Pittsburgh, PA, USA). Primary PCR reactions were performed using primers specific to the 5' UTR and 3' UTR (Table S1). The PCR products were re-amplified using primers that spanned all possible exon pairs. PCR products were separated on an agarose gel, visualized with ethidium bromide and analyzed using an ImageQuant LAS-4000 gel imager.
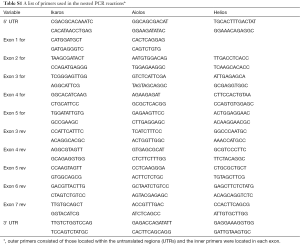
Full table
Statistics
For qRT-PCR data, statistics were performed using the one-way ANOVA with a Tukey posthoc test, and significance was defined as P<0.05. For linear regression, whether the slope of the line was different than zero was determined and significance was defined as P<0.05. Statistical analysis was performed using GraphPad Prism (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Cells from T-ALL patients with a CD3− DN phenotype
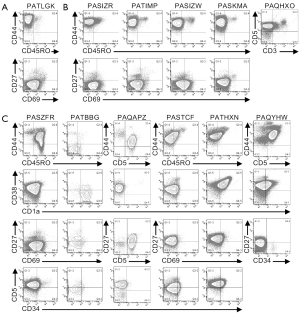
Multi-parameter flow cytometry was used to define the phenotypic characteristics of the cells from pediatric T-ALL patients (Figures 1,2,S1,S2). Cells from one patient (PATLGK) were mostly CD7− and were CD3−CD4−CD8−CD34+CD1a− (Figure 1A), suggesting that they arose from an early developmental stage, possibly early thymic progenitors (36). However, these cells also expressed CD38, which is not typically observed until the DN2 developmental stage (3,4).
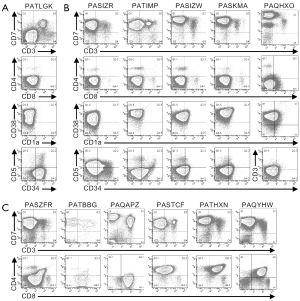
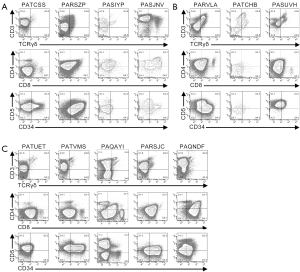
Cells from five T-ALL patients were CD7+ and CD3−CD4−CD8− (Figure 1B). In each of these five patients, the majority of the cells were CD38+CD1a−, but the expression of CD38 and CD1a varied slightly among the patients. For example, cells from patient PASIZR were CD38lo and resembled a DN1/DN2 transition state. By contrast, cells from patients PASIZW and PASKMA were CD1alo, consistent with a DN2/DN3 transition. Most CD3−CD4−CD8− T-ALL cells were CD44hiCD27−CD45RO−, but had varying levels of CD5 and CD69 (Figure S1A,B). In normal human thymocytes, CD5 is expressed on almost all cells and CD69 is not expressed until after cells express surface CD3 (37-39).
Pediatric T-ALL cells that lacked CD3, but expressed either CD4 or CD8
Six CD7+CD3− leukemia samples expressed varying levels of CD4 or CD8 (Figure 1C). We classified patient PAQAPZ as CD3− because the CD3− cells were CD8lo and were likely the leukemic cells while the CD3+ cells resembled normal peripheral T cells. Cells from three patients (PASTCF, PATHXN, and PAQYHW) were predominantly CD4+ and the CD8 levels ranged from negative to high, phenotypes consistent with the ISP CD4+ and DP developmental stages. Three patients (PASZFR, PATBBG, and PAQAPZ) were CD3−CD4− and expressed low levels of CD8, a phenotype that is rare among normal thymocytes (40).
Among the CD7+CD3− leukemia samples, cells from patient PATBBG were unique in that they lacked CD44, CD45RO, and CD38 expression on their surface (Figure S1C). Cells from the three patients that expressed ISP CD4+ and DP markers had varying levels of CD45RO with the DP-like T-ALL cells expressing the highest levels of CD45RO (Figure S1C); during normal T cell development, increased CD45RO expression correlates with progression to the DP developmental stage (41-43). Cells from patient PASZFR, which were CD3−CD4−CD8lo, were also CD45RO+ and had low levels of CD69. CD38 and CD1a expression was highly variable across the six samples and did not correlate with CD4 and CD8 expression. Cells from five of the six patients expressed CD27, even though CD27 is not normally expressed until the late stages of T cell development (39).
CD3+ pediatric T-ALL cells
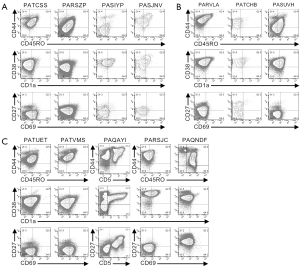
Cells from the remaining twelve patients expressed CD3 on their surface (Figures 2,S2). Cells from four patients (PATCSS, PARSZP, PASIYP, and PASJNV) were CD4−CD8−, CD4+CD8−/lo, or CD4+CD8+ (Figure 2A). Cells from each of these four samples expressed CD45RO and CD38, but variable levels of CD44 and CD1a (Figure S2A). Three of these samples expressed low levels of CD27. Cells from patients PARSZP and PASJNV also expressed CD34.
Cells from three patients (PARVLA, PATCHB, and PASUVH) expressed TCRγδ (Figure 2B). Cells from patient PATCHB lacked CD4 and CD8 expression, but the other two samples expressed CD4 and cells from patient PARVLA also expressed low levels of CD8. Cells from each of these three patients were primarily CD38+CD1aloCD69− (Figure S2B). Expression of CD44 and CD27 varied among these patients.
Cells from five patients were CD3+TCRγδ− and primarily CD8loCD4− (Figure S2C). Cells from two of these patients (PATVMS and PARSJC) could be subdivided into CD34lo and CD34hi subsets while cells from patient PAQNDF were uniformly CD34lo. Cells from patients PATUET and PAQAYI were predominantly CD34−. Most cells from these six patients expressed CD44, CD45RO, CD38, and CD1a (Figure S2C). Cells from two patients expressed CD27 and three patients had low levels of CD69 expression.
In summary, the phenotyping data indicate that correlating the surface phenotype of the leukemic cells to normal thymocyte populations is rarely possible. In many cases, cells expressed combinations of markers that were inconsistent with any normal thymocyte populations. Further, none of the samples shared identical phenotypes.
mRNA levels of Ikaros family members vary among T-ALL cells
For each T-ALL sample, we determined the relative mRNA levels of each of the five Ikaros family members (Figure 3A). Samples were grouped according to their expression of CD4, CD8 and TCRδ and, where possible, the mRNA levels were compared to normal thymocytes within each phenotype. Of the seven T-ALL samples with a DN phenotype, Ikaros mRNA levels were elevated in five samples, Helios mRNA levels in four samples, Aiolos mRNA levels in five samples, and Pegasus mRNA levels in six samples, as compared to normal DN thymocytes. Two patients had low Ikaros mRNA levels, one had low Helios mRNA levels, one had low Aiolos mRNA levels, and six had low Eos mRNA levels.
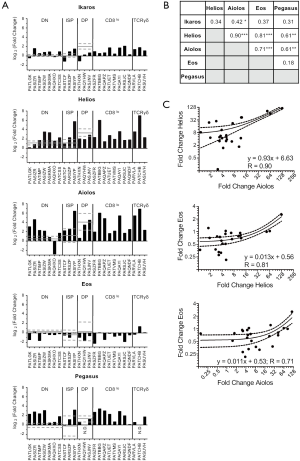
Among the three T-ALL samples with an ISP phenotype, one patient had elevated Ikaros, Helios, Aiolos, and Pegasus mRNA levels. One patient had elevated Ikaros and Helios mRNA levels. The third patient had low Ikaros mRNA levels. All three patients whose T-ALL cells were CD4+CD8+ had low Ikaros mRNA levels, as compared to normal DP thymocytes. Two of these patients had low Helios mRNA levels; one of these also had low Aiolos and Pegasus mRNA levels and the other had low Eos mRNA levels.
For patients whose cells were CD8lo or TCRγδ+, we compared the expression of each Ikaros family member across the samples. Helios and Aiolos mRNA levels varied most dramatically in these patients; the samples that expressed the highest Helios and Aiolos mRNA levels had 47-fold and 38-fold more than samples with the lowest mRNA levels, respectively. By contrast, cells expressing the highest levels of Ikaros, Eos, and Pegasus had 3.5-fold, 12-fold, and 10-fold more mRNA, respectively, than cells expressing the lowest levels of these family members.
Next, we used linear regression to determine whether the mRNA levels of any Ikaros family member correlated with that of any other family member (Figure 3B,C). There were strong correlations between the mRNA levels of Helios and Aiolos, Helios and Eos, and Aiolos and Eos. There were weak but statistically significant correlations between Helios and Pegasus and between Aiolos and Pegasus. Further, there was a very weak, but statistically significant correlation between Ikaros and Aiolos.
In conclusion, the mRNA levels of the Ikaros family members do not correlate with the surface phenotype of the T-ALL cells. However, the mRNA levels of Helios, Aiolos, and Eos correlated strongly with each other, suggesting that these genes are regulated similarly or these genes regulate each other.
The contribution of each Ikaros family member to the total Ikaros pool varies
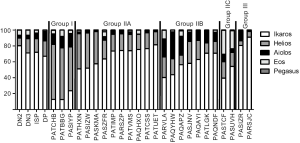
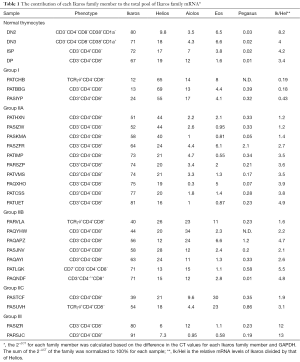
Next, we calculated the percentage of total Ikaros family mRNA represented by each family member (Figure 4 and Table 1). The majority (14/24) of T-ALL samples had a lower percentage of Ikaros family members represented by Ikaros than normal thymocytes. By contrast, Helios was overrepresented in 16/24 T-ALL samples, as compared to normal cells. Aiolos mRNA represented 1% or less of the total Ikaros mRNA in four T-ALL samples, but more than 20% of total Ikaros mRNA in three samples. Eos mRNA was underrepresented, as compared to normal thymocytes, in ten T-ALL samples. Pegasus represented less than 0.03% of total Ikaros mRNA in normal thymocytes, but more than 0.5% in four T-ALL samples and 2.1% in one case.
Full table
Next, we determined whether the T-ALL patients could be categorized based on the distribution of Ikaros mRNA levels. Because Ikaros and Helios accounted for most of the Ikaros family mRNA in nearly all T-ALL samples, we first calculated the ratio of Ikaros mRNA to Helios mRNA. Three groups of patients emerged, as shown in Table 1. Group I included samples in which the ratio of Ikaros to Helios mRNA levels was less than 0.5. Group II samples included those with an Ikaros to Helios ratio between 0.5 and 10. Group III included samples in which the Ikaros to Helios ratio exceeded 10. Group II could be subdivided into three subgroups based on the expression of Aiolos and Eos. Group IIA samples had minimal contribution from either Aiolos or Eos. Aiolos accounted for more than 10% of the total Ikaros mRNA in samples from group IIB and Eos accounted for more than 20% of total Ikaros in samples in group IIC.
The categorization of patients based on Ikaros family expression was independent of the surface phenotype of the cells. For example, most T-ALL groups included patients whose cells were CD3− and other patients whose cells were CD3+.
Protein levels and alternative splicing of Ikaros, Aiolos, and Helios in T-ALL
We used western blot analysis to examine Ikaros and Aiolos protein levels and splicing in cells from each T-ALL patient and in normal DN thymocytes (Figure 5). Ikaros protein levels in one patient were similar to that of normal DN thymocytes, but the Ikaros levels were less than half of normal in the remaining samples. The patient with the highest Ikaros protein levels, PAQNDF, had lower Ikaros mRNA levels than 18 of the other 23 samples. Similarly, most T-ALL samples expressed less full-length Aiolos protein than normal DN thymocytes.
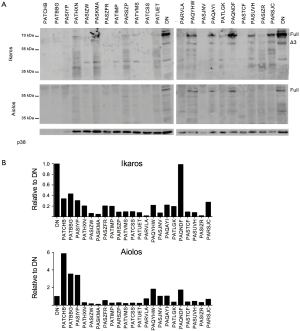
Low molecular weight bands were detected when membranes were probed with anti-Ikaros and anti-Aiolos, suggestive of the presence of alternative splice variants. We performed nested PCR using the primer pairs illustrated in Figure 6A to determine whether Ikaros, Helios, and Aiolos might undergo alternative splicing in normal thymocytes and T-ALL cells. A complex pattern of Ikaros splice variants was detected in normal and leukemic cells, consistent with our previous observations (40) and that different isoforms can lack exons, lack portions of exons, or include intronic sequences (19,20,44). In contrast to Ikaros, few T-ALL samples expressed multiple splice variants of Aiolos and Helios.
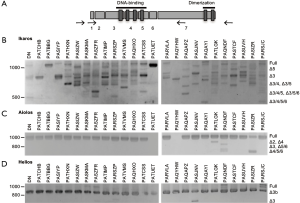
To verify the identity of the splice variants, nested RT-PCR was performed using primers that span each combination of exons (a sample of which is shown in Figure S3). For Ikaros, the most abundant proteins detected by western blot are most likely full-length Ikaros and Ikaros lacking exon 3 (Ik-∆3). In addition, numerous other splice variants were detected at the mRNA level; the most prevalent of which are shown in Figure 6B.
For Aiolos, full-length mRNA was detected in most samples (Figure 6C). Other splice variants detected lacked exon 2 (Ai-∆2), exon 3 (Ai-∆3), or exon 4 (Ai-∆4) or a combination of two or more exon deletions, such as Ai-∆5/6 and Ai-∆4/5/6. Helios lacking a portion of exon 3 (Hel-∆3b) was the most evident splice variant observed (Figure 6D), but some samples expressed full-length Helios and Helios lacking the entire third exon (Hel-∆3).
Based on these data, we conclude that the mRNA and protein levels of Ikaros and Aiolos do not directly correlate, which is consistent with our previous observations in normal human thymocytes (40). Further, many splice variants are not translated at detectable levels. For most splice variants detected, the isoforms also exist in normal thymocytes.
Discussion
The multi-parameter phenotypic analysis of cells from pediatric T-ALL patients revealed great heterogeneity among patients. Even when analyzing a limited number of markers, leukemic cells resembled normal human thymocyte populations in only nine of the 24 samples studied. For the majority of the samples, the phenotype deviated significantly from that of known normal human thymocyte populations. For example, cells from some T-ALL patients were CD8lo with little or no CD4 expression. The CD3−CD4−CD8lo T-ALL cells are similar to the ISP population observed during murine T cell development and a population of human thymocytes found in a fraction of patients (Mitchell et al., unpublished data). The CD3+CD8lo leukemia cells expressed lower levels of CD8 than mature SP CD8+ thymocytes and they did not express CD69 or CD27, making it unlikely that these are mature thymocytes. Among the T-ALL samples were those in which the cells expressed markers representing conflicting stages of development. For example, some cells co-expressed CD3 and CD34; in normal thymocytes, CD34 expression is terminated prior to CD3 expression.
Collectively, our data indicate that correlating T-ALL cells to normal developmental stages is rarely possible. In addition, each T-ALL sample had a unique phenotype. These two features make classifying T-ALL patients a challenging endeavor, as seen by the difficulty in using phenotype as a prognostic indicator (11,12,45).
As an alternate method to classify T-ALL patients, we focused on the Ikaros family of transcription factors, which was previously implicated in leukemogenesis. ChIP-seq data showed that the Ikaros family can bind 4,500–5,600 sites in the genome and regulate several hundred genes (46,47), making these transcription factors key regulators of cell fate decisions in developing lymphocytes. The importance of the Ikaros family in leukemogenesis has been most clearly demonstrated in mice, where expression of dominant negative isoforms of Ikaros or Helios resulted in T cell malignancies (14-16,18). Disease onset in these mice was dependent on the reduced functionality of the entire Ikaros family, as deletion of individual family members did not cause disease (17,48,49). This observation highlights the importance of studying the entire family. In humans, correlative studies have suggested the presence of dominant negative isoforms of Ikaros in T-ALL patients, but the percentage of patients harboring these splice variants is controversial (19-22,27,44,50,51). Our data indicate that alternative mRNA splicing of Ikaros is common in normal thymocytes and T-ALL cells, but not all splice variants are translated into protein products (Figures 5,6).
The mRNA levels of each Ikaros family member varied widely across T-ALL samples, but the greatest disparity was in Aiolos and Helios expression (Figure 3A). The samples with the highest Aiolos and Helios mRNA levels were 780-fold and 120-fold greater, respectively, than the samples with the lowest mRNA levels. This level of variability is in contrast to the 18-fold and 10-fold changes in Aiolos and Helios mRNA levels, respectively, that normally occur as thymocytes progress from the DN to the DP stages. By contrast, samples with the highest mRNA levels of Ikaros, Eos, and Pegasus only contained 8.6- to 11-fold more than the samples with the lowest levels.
Based on these data, we propose the classification of T-ALL patients using the strategy shown in Figure 4 and Table 1. For most patients, Ikaros and Helios mRNA was the primary Ikaros family member transcripts detected, so the ratio of Ikaros mRNA levels to Helios mRNA levels was used to divide the samples into three groups. The largest group of patients had Ikaros to Helios ratios similar to those of normal thymocytes. Within this group, the mRNA levels of Aiolos and Eos varied in a manner that provided a means to subdivide these patients.
The importance of Ikaros family mRNA levels in leukemogenesis may arise from the function of the Ikaros family proteins. Because of the limited number of cells we could obtain from the patients, we were only able to analyze Ikaros and Aiolos protein levels. For both proteins, the protein levels did not correlate with mRNA levels and the protein levels were lower than in normal thymocytes for most patients (Figures 3,5). The reduction of Ikaros family proteins may be linked to leukemogenesis, as it is in mice (14-18,48,49).
In addition to a general reduction of Ikaros family expression, the ratio of Ikaros to Aiolos protein varied across the T-ALL samples, suggesting the dimer composition may vary among patients. For example, patient PATCHB had high Aiolos levels and low Ikaros levels whereas patient PAQNDF had the opposite pattern. Even small changes in the ratio among Ikaros family members can have profound biological effects, as seen by the development of lymphoma when Helios was expressed at low levels in murine B cells (17).
While Ikaros family protein levels have direct implications in leukemogenesis, the mechanisms that control Ikaros family transcription might control other genes that regulate the development of T-ALL. For example, c-myc, IRF4, Runx1, and TAL-1 can regulate Ikaros expression and are activated in some T-ALL patients (52-61). In addition, there are Ikaros binding sites within the promoters of Ikaros, Aiolos, Eos, and Pegasus (54,62) and the correlations in the expression of some Ikaros family members we observed (Figure 3) supports a model in which Ikaros family members may regulate the transcription of themselves and other family members. Further, the Ikaros binding site is shared by all family members and by the Notch-dependent transcription factor RBP-Jκ (63-66). Because half of pediatric T-ALL patients have an activating mutation in Notch (67), it would be expected that Ikaros family mRNA levels would be altered in these patients.
In summary, we used multi-parameter flow cytometry to demonstrate the tremendous variability among pediatric T-ALL samples. Further, we performed the first comprehensive analysis of the expression of the entire Ikaros family of transcription factors reported in pediatric T-ALL. More research is needed to extend these observations and determine whether the patterns of Ikaros mRNA or protein levels correlate to clinical outcomes.
Acknowledgements
The authors would like to thank Dr. Keith August for his assistance in obtaining T-ALL samples. The authors would also like to thank Dr. Robert H. Ardinger Jr, Dr. James E. O’Brien Jr, Jennifer Marshall, and Diana Connelly for their assistance in obtaining the human thymus samples. In addition, the authors would like to thank Drs. Steve Benedict, Marci Chan, Mary Markiewicz, and members of their laboratories for helpful discussions.
Funding: This work was supported, in part, by the American Cancer Society Research Scholar Grant 08-182-LIB and the University of Kansas Cancer Center Pilot Grant. We acknowledge the Flow Cytometry Core Laboratory, which is sponsored, in part, by the NIH/NIGMS COBRE grant P30 GM103326. JL Mitchell was supported by a Madison and Lila Self Graduate Fellowship. We also acknowledge Chair’s Grant U10 CA98543 and Human Specimen Banking Grant U24 CA 114766 of the Children’s Oncology Group from the National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the Institutional Review Boards at Children’s Mercy Hospital and the University of Kansas Medical Center and written informed consent was obtained from all parent or guardian.
References
- Asnafi V, Beldjord K, Boulanger E, et al. Analysis of TCR, pT alpha, and RAG-1 in T-acute lymphoblastic leukemias improves understanding of early human T-lymphoid lineage commitment. Blood 2003;101:2693-703. [Crossref] [PubMed]
- Bene MC, Castoldi G, Knapp W, et al. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL). Leukemia 1995;9:1783-6. [PubMed]
- Staal FJ, Weerkamp F, Langerak AW, et al. Transcriptional control of t lymphocyte differentiation. Stem Cells 2001;19:165-79. [Crossref] [PubMed]
- Dik WA, Pike-Overzet K, Weerkamp F, et al. New insights on human T cell development by quantitative T cell receptor gene rearrangement studies and gene expression profiling. J Exp Med 2005;201:1715-23. [Crossref] [PubMed]
- Van de Walle I, De Smet G, De Smedt M, et al. An early decrease in Notch activation is required for human TCR-alphabeta lineage differentiation at the expense of TCR-gammadelta T cells. Blood 2009;113:2988-98. [Crossref] [PubMed]
- García-Peydró M, de Yebenes VG, Toribio ML. Sustained Notch1 signaling instructs the earliest human intrathymic precursors to adopt a gammadelta T-cell fate in fetal thymus organ culture. Blood 2003;102:2444-51. [Crossref] [PubMed]
- Taghon T, Van de Walle I, De Smet G, et al. Notch signaling is required for proliferation but not for differentiation at a well-defined beta-selection checkpoint during human T-cell development. Blood 2009;113:3254-63. [Crossref] [PubMed]
- Carrasco YR, Trigueros C, Ramiro AR, et al. Beta-selection is associated with the onset of CD8beta chain expression on CD4(+)CD8alphaalpha(+) pre-T cells during human intrathymic development. Blood 1999;94:3491-8. [PubMed]
- Takeuchi Y, Fujii Y, Okumura M, et al. Characterization of CD4+ single positive cells that lack CD3 in the human thymus. Cell Immunol 1993;151:481-90. [Crossref] [PubMed]
- Asnafi V, Buzyn A, Thomas X, et al. Impact of TCR status and genotype on outcome in adult T-cell acute lymphoblastic leukemia: a LALA-94 study. Blood 2005;105:3072-8. [Crossref] [PubMed]
- van Grotel M, van den Heuvel-Eibrink MM, van Wering ER, et al. CD34 expression is associated with poor survival in pediatric T-cell acute lymphoblastic leukemia. Pediatr Blood Cancer 2008;51:737-40. [Crossref] [PubMed]
- Pui CH, Hancock ML, Head DR, et al. Clinical significance of CD34 expression in childhood acute lymphoblastic leukemia. Blood 1993;82:889-94. [PubMed]
- van Grotel M, Meijerink JP, van Wering ER, et al. Prognostic significance of molecular-cytogenetic abnormalities in pediatric T-ALL is not explained by immunophenotypic differences. Leukemia 2008;22:124-31. [Crossref] [PubMed]
- Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell 1995;83:289-99. [Crossref] [PubMed]
- Papathanasiou P, Perkins AC, Cobb BS, et al. Widespread failure of hematolymphoid differentiation caused by a recessive niche-filling allele of the Ikaros transcription factor. Immunity 2003;19:131-44. [Crossref] [PubMed]
- Kirstetter P, Thomas M, Dierich A, et al. Ikaros is critical for B cell differentiation and function. Eur J Immunol 2002;32:720-30. [Crossref] [PubMed]
- Dovat S, Montecino-Rodriguez E, Schuman V, et al. Transgenic expression of Helios in B lineage cells alters B cell properties and promotes lymphomagenesis. J Immunol 2005;175:3508-15. [Crossref] [PubMed]
- Zhang Z, Swindle CS, Bates JT, et al. Expression of a non-DNA-binding isoform of Helios induces T-cell lymphoma in mice. Blood 2007;109:2190-7. [Crossref] [PubMed]
- Sun L, Crotty ML, Sensel M, et al. Expression of dominant-negative Ikaros isoforms in T-cell acute lymphoblastic leukemia. Clin Cancer Res 1999;5:2112-20. [PubMed]
- Sun L, Heerema N, Crotty L, et al. Expression of dominant-negative and mutant isoforms of the antileukemic transcription factor Ikaros in infant acute lymphoblastic leukemia. Proc Natl Acad Sci U S A 1999;96:680-5. [Crossref] [PubMed]
- Nakase K, Ishimaru F, Avitahl N, et al. Dominant negative isoform of the Ikaros gene in patients with adult B-cell acute lymphoblastic leukemia. Cancer Res 2000;60:4062-5. [PubMed]
- Ruiz A, Jiang J, Kempski H, et al. Overexpression of the Ikaros 6 isoform is restricted to t(4;11) acute lymphoblastic leukaemia in children and infants and has a role in B-cell survival. Br J Haematol 2004;125:31-7. [Crossref] [PubMed]
- Kuiper RP, Schoenmakers EF, van Reijmersdal SV, et al. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia 2007;21:1258-66. [Crossref] [PubMed]
- Maser RS, Choudhury B, Campbell PJ, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature 2007;447:966-71. [Crossref] [PubMed]
- Mullighan CG, Miller CB, Radtke I, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 2008;453:110-4. [Crossref] [PubMed]
- Meleshko AN, Movchan LV, Belevtsev MV, et al. Relative expression of different Ikaros isoforms in childhood acute leukemia. Blood Cells Mol Dis 2008;41:278-83. [Crossref] [PubMed]
- Marçais A, Jeannet R, Hernandez L, et al. Genetic inactivation of Ikaros is a rare event in human T-ALL. Leuk Res 2010;34:426-9. [Crossref] [PubMed]
- Hahm K, Cobb BS, McCarty AS, et al. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev 1998;12:782-96. [Crossref] [PubMed]
- Kelley CM, Ikeda T, Koipally J, et al. Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr Biol 1998;8:508-15. [Crossref] [PubMed]
- Morgan B, Sun L, Avitahl N, et al. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J 1997;16:2004-13. [Crossref] [PubMed]
- Perdomo J, Holmes M, Chong B, et al. Eos and pegasus, two members of the Ikaros family of proteins with distinct DNA binding activities. J Biol Chem 2000;275:38347-54. [Crossref] [PubMed]
- Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J 1996;15:5358-69. [PubMed]
- Schjerven H, McLaughlin J, Arenzana TL, et al. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat Immunol 2013;14:1073-83. [Crossref] [PubMed]
- Molnár A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol 1994;14:8292-303. [Crossref] [PubMed]
- Reyes-León A, Juarez-Velazquez R, Medrano-Hernandez A, et al. Expression of Ik6 and Ik8 Isoforms and Their Association with Relapse and Death in Mexican Children with Acute Lymphoblastic Leukemia. PloS One 2015;10:e0130756. [Crossref] [PubMed]
- Chakraverty R, Flutter B, Fallah-Arani F, et al. The host environment regulates the function of CD8+ graft-versus-host-reactive effector cells. J Immunol 2008;181:6820-8. [Crossref] [PubMed]
- Awong G, Herer E, Surh CD, et al. Characterization in vitro and engraftment potential in vivo of human progenitor T cells generated from hematopoietic stem cells. Blood 2009;114:972-82. [Crossref] [PubMed]
- Jung LK, Haynes BF, Nakamura S, et al. Expression of early activation antigen (CD69) during human thymic development. Clin Exp Immunol 1990;81:466-74. [Crossref] [PubMed]
- Vanhecke D, Leclercq G, Plum J, et al. Characterization of distinct stages during the differentiation of human CD69+CD3+ thymocytes and identification of thymic emigrants. J Immunol 1995;155:1862-72. [PubMed]
- Mitchell JL, Seng A, Yankee TM. Ikaros, Helios, and Aiolos protein levels increase in human thymocytes after β selection. Immunol Res 2016;64:565-75. [Crossref] [PubMed]
- Janossy G, Bofill M, Rowe D, et al. The tissue distribution of T lymphocytes expressing different CD45 polypeptides. Immunology 1989;66:517-25. [PubMed]
- Gillitzer R, Pilarski LM. In situ localization of CD45 isoforms in the human thymus indicates a medullary location for the thymic generative lineage. J Immunol 1990;144:66-74. [PubMed]
- Okumura M, Fujii Y, Inada K, et al. CD45RA-R0+ subset is the major population of dividing thymocytes in the human. Eur J Immunol 1992;22:3033-6. [Crossref] [PubMed]
- Sun L, Goodman PA, Wood CM, et al. Expression of aberrantly spliced oncogenic ikaros isoforms in childhood acute lymphoblastic leukemia. J Clin Oncol 1999;17:3753-66. [PubMed]
- Sidhom I, Shaaban K, Soliman S, et al. Clinical significance of immunophenotypic markers in pediatric T-cell acute lymphoblastic leukemia. J Egypt Natl Canc Inst 2008;20:111-20. [PubMed]
- Ferreirós-Vidal I, Carroll T, Taylor B, et al. Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood 2013;121:1769-82. [Crossref] [PubMed]
- Geimer Le Lay AS, Oravecz A, Mastio J, et al. The tumor suppressor Ikaros shapes the repertoire of notch target genes in T cells. Sci Signal 2014;7:ra28. [Crossref] [PubMed]
- Wang JH, Nichogiannopoulou A, Wu L, et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 1996;5:537-49. [Crossref] [PubMed]
- Wang JH, Avitahl N, Cariappa A, et al. Aiolos regulates B cell activation and maturation to effector state. Immunity 1998;9:543-53. [Crossref] [PubMed]
- Klein F, Feldhahn N, Herzog S, et al. BCR-ABL1 induces aberrant splicing of IKAROS and lineage infidelity in pre-B lymphoblastic leukemia cells. Oncogene 2006;25:1118-24. [Crossref] [PubMed]
- Nückel H, Frey UH, Sellmann L, et al. The IKZF3 (Aiolos) transcription factor is highly upregulated and inversely correlated with clinical progression in chronic lymphocytic leukaemia. Br J Haematol 2009;144:268-70. [Crossref] [PubMed]
- Palomero T, Lim WK, Odom DT, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A 2006;103:18261-6. [Crossref] [PubMed]
- Margolin AA, Palomero T, Sumazin P, et al. ChIP-on-chip significance analysis reveals large-scale binding and regulation by human transcription factor oncogenes. Proc Natl Acad Sci U S A 2009;106:244-9. [Crossref] [PubMed]
- Yoshida T, Landhuis E, Dose M, et al. Transcriptional regulation of the Ikzf1 locus. Blood 2013;122:3149-59. [Crossref] [PubMed]
- La Starza R, Borga C, Barba G, et al. Genetic profile of T-cell acute lymphoblastic leukemias with MYC translocations. Blood 2014;124:3577-82. [Crossref] [PubMed]
- Adamaki M, Lambrou GI, Athanasiadou A, et al. Implication of IRF4 aberrant gene expression in the acute leukemias of childhood. PloS One 2013;8:e72326. [Crossref] [PubMed]
- Grossmann V, Kern W, Harbich S, et al. Prognostic relevance of RUNX1 mutations in T-cell acute lymphoblastic leukemia. Haematologica 2011;96:1874-7. [Crossref] [PubMed]
- Della Gatta G, Palomero T, Perez-Garcia A, et al. Reverse engineering of TLX oncogenic transcriptional networks identifies RUNX1 as tumor suppressor in T-ALL. Nat Med 2012;18:436-40. [Crossref] [PubMed]
- Ma S, Pathak S, Trinh L, et al. Interferon regulatory factors 4 and 8 induce the expression of Ikaros and Aiolos to down-regulate pre-B-cell receptor and promote cell-cycle withdrawal in pre-B-cell development. Blood 2008;111:1396-403. [Crossref] [PubMed]
- Ferrando AA, Neuberg DS, Staunton J, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell 2002;1:75-87. [Crossref] [PubMed]
- Brown L, Cheng JT, Chen Q, et al. Site-specific recombination of the tal-1 gene is a common occurrence in human T cell leukemia. EMBO J 1990;9:3343-51. [PubMed]
- Ghadiri A, Duhamel M, Fleischer A, et al. Critical function of Ikaros in controlling Aiolos gene expression. FEBS Lett 2007;581:1605-16. [Crossref] [PubMed]
- Beverly LJ, Capobianco AJ. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell 2003;3:551-64. [Crossref] [PubMed]
- Kathrein KL, Chari S, Winandy S. Ikaros directly represses the notch target gene Hes1 in a leukemia T cell line: implications for CD4 regulation. J Biol Chem 2008;283:10476-84. [Crossref] [PubMed]
- Chari S, Winandy S. Ikaros regulates Notch target gene expression in developing thymocytes. J Immunol 2008;181:6265-74. [Crossref] [PubMed]
- Kleinmann E, Geimer Le Lay AS, Sellars M, et al. Ikaros represses the transcriptional response to Notch signaling in T-cell development. Mol Cell Biol 2008;28:7465-75. [Crossref] [PubMed]
- Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004;306:269-71. [Crossref] [PubMed]