Meta-signature of mutated genes in gallbladder cancer: evidence based high throughput screening assays
Background
Gallbladder carcinoma (GBC) is the fifth most common carcinoma of gastrointestinal tract, and represents 80–95% of biliary tract cancers. It is relatively an uncommon malignant disease with a poor prognosis. According to previous reports (1), GBC has a low incidence rate (<2/100,000). Reid et al. (2) found that the worldwide incidence of GBC correlates with the prevalence of gallstone disease. The high-incidence areas of GBC are Poland (14/100,000), Northern India (21.5/100,000), south Pakistan (11.3/100,000), Israel (5/100,000) and Japan (7/100,000) (1). Besides, GBC is more common in females. Stinton et al. (1) demonstrated that the incidence rate was high in South American females, 15.5 per 100,000 in Bolivia (vs. 7.5/100,000 in male), and 11.3 per 100,000 in New Mexico (vs. 4/100,000 in male).
A satisfied outcome depends on the early diagnosis and appropriative treatment. Up to date, the most effective treatment for GBC patients is surgery. However, mainly due to their occult symptoms, less than 10% of GBC patients have the opportunities to receive surgery, and nearly 50% of them already had lymph node metastasis at first diagnosis. Because of the difficulties in early diagnosis, the prognosis of GBC is so poor. The overall 5-year survival rate of GBC patients is less than 5% (3). A thorough understanding of the underlying mechanism is critical for exploring potential diagnostic biomarker and developing effective therapeutic approach for GBC patients.
High-throughput genetic mutation profiling in GBC
Grateful thanks to the decades of relevant studies, a numerous molecular mechanisms involved in GBC were unveiled. Recently, molecular testing in multiple solid tumors has become standard practice. Newer molecular tests are focusing on mutation detection as a diagnostic biomarker of GBC. High-throughput genetic mutation profiling provided the possibility to do the comprehensive examination of the cancer genome. It has undoubted advances in the characterization and quantification of genomes, epigenomes and transcriptomes. High-throughput genetic mutation profiling is being widely applied in mutation detection. Today, several commercial platforms are available, including SNaPshot multiplex system, next generation sequencing (NGS) and massARRAY platform technics. Among of them, NGS technology is widely applied high-throughput genetic mutation detection method since 2006. NGS technology is free from many of the confines dictated by previous technologies, such as the bias due to the probe selection in array technology, cross-hybridization background, and signal saturation-induced detection dynamic range limitation.
Recently, Javle et al. (4) performed mass spectroscopy-based and next-generation sequencing profiling in GBC samples. By hotspot mutations analysis, they found 14 hotspot mutations from 11 different genes, included IDH1, KRAS, NRAS, PIK3CA and MET. Among of them, mutations in IDH1 are the most recurrent (36.4%). They also detected 26 mutations by targeted NGS, and identified TP53 as the most common mutated gene. They further conducted a multivariate analysis and found mutated IDH and KRAS were associated with poorer overall survival. Their results provided evidence that high-throughput mutation profiling may be a useful platform for identifying novel mutations for targeted therapy of GBC.
Meta-signature of mutated genes in GBC
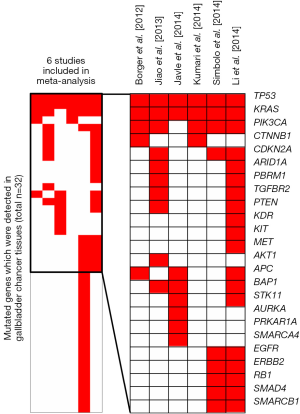
Nowadays, increasing groups are focusing on mutated genes in GBC. However, due to small sample size and different technological platforms between above studies, the mutated gene profiling effort in GBC led to inconsistent results. To overcome the limitations, we conducted a meta-signature of mutated genes in GBC based on six studies (4-9) including 232 subjects receiving high-throughput genetic mutation profiling (Table 1). Totally 43 mutated genes were detected in 232 GBC patients. Among of them, six genes (TP53, KRAS, PIK3CA, CDKN2A, BAP1 and APC) were reported in more than three studies (Figure 1). Our meta-analysis further revealed that three mutated genes (TP53, KRAS, PIK3CA) were significantly associated with GBC (Table 2). In the following aspect, we will discuss the three recurrent mutated genes.
Full table

Full table
TP53 contains 34,453 mutations, including 1,311 hot-spot mutations (10). Increasing evidence suggest that mutated TP53 plays important role in multiple tumors. Cardesa et al. (11) represented that TP53 gene mutations were observed in up to 50% of head and neck squamous-cell carcinomas and approximately 65% of them have aberrant expression of TP53. Szymańska et al. (12) also reported that TP53 was the most frequently mutated gene in human cancer, such as hepatocellular carcinoma and oesophagus carcinoma. Asai et al. (13), exploredTP53 mutations in GBC patients, and found nearly half of GBC patients have TP53 mutations. In our meta-analysis, we also found that TP53 was the most recurrent mutated gene in GBC (crude P value =1.44×10–6, corrected P value =1.00×10–3, Table 2).
There are more than 3,000 in KRAS, and 90% of them are located in exon 2 and 10% in exons 3 and 4 (www.sanger.ac.uk/genetics/CGP/cosmic/). KRAS has been considered as one of the most frequently mutated genes in multiple tumors. Therkildsen et al. (14) meta-analyzed 22 studies with 2,395 patients with different tumors, and found that KRAS mutations might be implemented for prediction of clinical benefit from anti-EGFR antibodies in metastatic colorectal cancer. Eirini et al. (15) explored KRAS mutations in non-small-cell lung cancer patients, and represented that KRAS exon 2 mutation was observed in 18.89% (106/561) patients. Reid et al. (2) reported that KRAS mutations were associated with GBC in patients with anomalous junction of the pancreaticobiliary duct (AJPBD), suggesting that KRAS mutation might serve as a useful tool in screening early GBC in patients with AJPBD. Our data also revealed that mutated KRAS was associated with GBC (crude P value =3.37×10–6, corrected P value =2.34×10–2, Table 2), consistent with previous studies.
PIK3CA is located on 3q26.3, whose mutations were also associated with multiple malignancies. Dey et al. (16) found that PIK3CA mutations were detected in 35% patients with breast cancer, which were associated with deregulation of PI3K pathway and contributed to carcinogenesis of breast cancer. Yip et al. (17) also reported the relationship between mutated PIK3CA and nasopharyngeal carcinoma (NPC). They performed qRT-PCR and immunohistochemical staining in 74 patients with NPC, and demonstrated that aberrant expression of PIK3CA was detected in 68.9% (51/74) patients with NPC. In GBC, Deshpande et al. (18) found PIK3CA mutations in 12.5% patients and suggested PIK3CA mutations as diagnostic biomarkers and therapy targets. In the present study, we also found that mutated PIK3CA was associated with GBC, although the corrected P-value was not significant mainly due to small number of studies (crude P value =1.10×10–4, corrected P value =7.65×10–2, Table 2).
Summary and prospect
Overall, our meta-analysis data strongly suggested that mutated TP53, KRAS, PIK3CA were associated with GBC, and it may be a potential diagnostic and prognostic biomarker for GBC patients. However, nowadays, the limited number of studies cannot supply sufficient evidence for further analysis. Therefore, large, multi-center and well-performed studies are warranted to confirm above findings. In future, GBC patients harboring mutations of TP53, KRAS, PIK3CA may benefit from target therapies available or in development.
Acknowledgements
Funding: This study was supported by National Science Foundation of China (No. 81071876, 81201549, 81272644 and 81472247), and the Project of Innovative Research for Key Science and Technology in Xi’an Jiaotong University (2013KCJ-23).
Footnote
Provenance: This is a Guest Commentary commissioned by Guest Editor Haitao Zhao, MD, PhD (Department of Liver Surgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China).
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Stinton LM, Shaffer EA. Epidemiology of gallbladder disease: cholelithiasis and cancer. Gut Liver 2012;6:172-87. [Crossref] [PubMed]
- Reid KM, Ramos-De la Medina A, Donohue JH. Diagnosis and surgical management of gallbladder cancer: a review. J Gastrointest Surg 2007;11:671-81. [Crossref] [PubMed]
- Rakić M, Patrlj L, Kopljar M, et al. Gallbladder cancer. Hepatobiliary Surg Nutr 2014;3:221-6. [PubMed]
- Javle M, Rashid A, Churi C, et al. Molecular characterization of gallbladder cancer using somatic mutation profiling. Hum Pathol 2014;45:701-8. [Crossref] [PubMed]
- Borger DR, Tanabe KK, Fan KC, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 2012;17:72-9. [Crossref] [PubMed]
- Jiao Y, Pawlik TM, Anders RA, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet 2013;45:1470-3. [Crossref] [PubMed]
- Kumari N, Corless CL, Warrick A, et al. Mutation profiling in gallbladder cancer in Indian population. Indian J Pathol Microbiol 2014;57:9-12. [Crossref] [PubMed]
- Simbolo M, Fassan M, Ruzzenente A, et al. Multigene mutational profiling of cholangiocarcinomas identifies actionable molecular subgroups. Oncotarget 2014;5:2839-52. [Crossref] [PubMed]
- Li M, Zhang Z, Li X, et al. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat Genet 2014;46:872-6. [Crossref] [PubMed]
- Edlund K, Larsson O, Ameur A, et al. Data-driven unbiased curation of the TP53 tumor suppressor gene mutation database and validation by ultradeep sequencing of human tumors. Proc Natl Acad Sci U S A 2012;109:9551-6. [Crossref] [PubMed]
- Cardesa A, Nadal A. Carcinoma of the head and neck in the HPV era. Acta Dermatovenerol Alp Pannonica Adriat 2011;20:161-73. [PubMed]
- Szymańska K, Hainaut P. TP53 and mutations in human cancer. Acta Biochim Pol 2003;50:231-8. [PubMed]
- Asai T, Loza E, Roig GV, et al. High frequency of TP53 but not K-ras gene mutations in Bolivian patients with gallbladder cancer. Asian Pac J Cancer Prev 2014;15:5449-54. [Crossref] [PubMed]
- Therkildsen C, Bergmann TK, Henrichsen-Schnack T, et al. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis. Acta Oncol 2014;53:852-64. [Crossref] [PubMed]
- Papadopoulou E, Tsoulos N, Tsirigoti A, et al. Determination of EGFR and KRAS mutational status in Greek non-small-cell lung cancer patients. Oncol Lett 2015;10:2176-84. [PubMed]
- Dey N, Leyland-Jones B, De P. MYC-xing it up with PIK3CA mutation and resistance to PI3K inhibitors: summit of two giants in breast cancers. Am J Cancer Res 2014;5:1-19. [PubMed]
- Yip WK, He PY, Abdullah MA, et al. Increased Expression of Phosphatidylinositol 3-Kinase p110α and Gene Amplification of PIK3CA in Nasopharyngeal Carcinoma. Pathol Oncol Res 2016;22:413-9. [Crossref] [PubMed]
- Deshpande V, Nduaguba A, Zimmerman SM, et al. Mutational profiling reveals PIK3CA mutations in gallbladder carcinoma. BMC Cancer 2011;11:60. [Crossref] [PubMed]