
Landscape of RB1 alterations in 22,432 Chinese solid tumor patients
Introduction
The human retinoblastoma susceptibility gene (RB1) is a tumor-suppressor gene is a member of a small gene family, to which RB11 (p107) and RB12 (p130) also belong. The RB1 protein (pRB) has important roles in multiple molecular processes, including gene transcription, DNA replication, DNA repair and mitosis (1). In an analysis of 932 databases, it was reported that pRB is often inactivated because of genetic nonsense and deletion mutations, with missense mutations being mainly responsible for loss of function in most cases (2).
As a chromatin-related protein, the RB1 protein plays an important role in regulating and controlling the cell cycle, acting as a component of the cyclin D-cyclin dependent kinase (CDK) 4/6-inhibitor of CDK4 (INK4A)/cyclin D1/pRB/E2 factor (E2F) regulatory pathway. In its active form, hypophosphorylated pRB suppresses the expression of genes associated with cell proliferation by binding E2F during the G1 phase of the cell cycle. At the G1/S transition, pRB is inactivated by phosphorylation, catalyzed by CDKs, leading to the disruption of E2F repressor complexes and the accumulation of activator E2F complexes that drive transcription, and finally allow cell-cycle progression (3). The regulation of the G1/S transition and cell proliferation are the most understood roles of RB1. The CDK/RB/E2F pathway is continually dysregulated in cancer cells, which leads to uncontrolled cell proliferation and progression. So far, there are 3 known CDK4/6 inhibitors, palbociclib, ribociclib, and abemaciclib. Despite the recent success of CDK inhibitors in hormone receptor (HR)-positive breast cancer, not all patients respond to these drugs. The loss of RB1 has been suggested to be one of the most important biomarkers for resistance to CDK4/6 inhibitors. Several previous preclinical and clinical research have also reported that RB1 mutations contribute to this resistance (4-8). Additionally, it is becoming clear that other roles, such as the regulation of the epithelial-to-mesenchymal transition (EMT) and potential influences on therapeutic effect to immune therapy (9,10), are also attributable to RB1, indicating that RB1 could be a novel treatment target for cancer therapy.
Cancer genome sequencing has revealed that the most common types of cancers with RB1 mutations are sarcomas, retinoblastomas, and SCLC, with mutation at lower frequencies in other cancer types, such as prostate cancer and breast cancer (3). However, to the best of our knowledge, no comprehensive analyses of the status of RB1 alterations in Chinese people with solid tumors have been reported. Therefore, it is necessary to fully characterize the alterations of RB1 in the Pan-Cancer background in this ethnicity. In the present study, we screened for molecular alterations of RB1 in 22,432 Chinese patients with a wide variety of solid tumors using next-generation sequencing (NGS) in a Clinical Laboratory Improvement Amendment (CLIA)-certified laboratory. The findings could indicate a new potential therapeutic target for the treatment of solid tumors, as well as provide a deeper understanding of RB1 for the treatment of cancer patients. Our data were extensive and comprehensive, with a large sample size of 21 different types of cancers, including non-small cell lung cancer (NSCLC), small cell lung cancer, colorectal cancer, and gastric cancer. We also analyzed the distribution of pathogenic RB1 (Retinoblastoma 1) mutations in hotspots and exons. In our study, we comprehensively described various types of RB1 mutation and mutation sites, which could serve as potential targets in drug development. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3162/rc).
Methods
Clinical specimens
We analyzed formalin-fixed, paraffin-embedded (FFPE) tissues from solid tumor patients who underwent NGS from January 1, 2017 to April 15, 2020 at the 3DMed Clinical Laboratory Inc. (a College of American Pathologists and CLIA-certified laboratory). Genomic DNA was isolated from tissue by using the ReliaPrep FFPE gDNA Miniprep System (Promega, Madison, Wisconsin, USA), quantified using the Qubit dsDNA HS assay kit (Thermo Fisher Scientific, USA) following the manufacturer’s specifications (11). Microsatellite instability/stability (MSI/MSS) status was assessed as follows: 100 microsatellite loci were selected for blood microsatellite instability determination; for each assay, the top 30 loci with the best coverage were included for the final MSI score calculation. The internally developed R script was used to evaluate the distribution of read counts between different repeat lengths of microsatellite site for each sample. The model to determine the stability of site was as follows:
where pi is the cumulative percentage of the MSS subtype cut-point repeat length (Ci), ni is number of unstable reads, and Ni is the number of reads for locus. If X ≥ ni, then it was considered unstable and the probability of P was ≤0.15%. The MSI score was defined as the proportion of unstable loci. The sample with an MSI score of ≥0.4 was defined as having high microsatellite instability (MSI-H), and the sample with an MSI score of <0.4 was defined as microsatellite stable (MSS).
NGS sequencing
Genomic DNA was processed by 3DMed Clinical Laboratory Inc for NGS using Illumina Nextseq 500 to >500× coverage, as previously described (12). The single-nucleotide variants (SNVs), copy number variants, and gene rearrangements data of these patients were analyzed. Clinical data were collected, including age, sex, and tumor histology. Each patient’s tumor tissue was compared with a matched blood control to determine whether germline variants were present.
Statistical analysis
Microsoft Excel (Microsoft, Redmond, WA, USA) was used in all of the analyses. Graphs included in the Figures 1-3 are visualized by Microsoft Excel 2013.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics committee of the First Affiliated Hospital of the Air Force Medical University (No. KY20222189-F-1), and informed consent was taken from all the patients.
Results
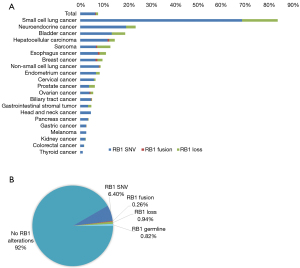
A total of 22,432 solid tumors from Chinese patients were included in the present study, consisting of 8,732 (38.9%) NSCLCs, 3,720 (16.6%) colorectal cancers, 1,649 (7.4%) hepatocellular carcinomas, 1,431 (6.4%) gastric cancers, 1,369 (6.1%) biliary tract cancers, and 910 (4.1%) pancreatic cancers, as well as other rarer histotypes (Table 1). The median age of the patients was 60 years, and 12,901 (57.5%) were male. RB1 mutations occurred in almost all solid tumors, with an overall mutation rate of 7.6% (1,712/22,432). RB1 alterations were more common among males than females (8.4% vs. 6.5%, P<0.001) (Table 1). RB1 mutations occurred most frequently in SCLC; 138/165, 83.6%), followed by neuroendocrine neoplasms (40/170, 23.5%), bladder cancer (40/209, 19.1%), hepatocellular carcinoma (233/1,649, 14.1%), sarcomas (71/554, 12.8%), and esophageal cancer (32/293, 10.9%).
Table 1
| Characteristics | All patient (n=22,432) | RB1 mutation | RB1 SNV | RB1 fusion | RB1 loss | RB1 germline mutation |
|---|---|---|---|---|---|---|
| Age (median) | 60 | |||||
| Sex, n (%) | ||||||
| Male | 12,901 (57.5) | 1,088 (8.4) | 939 (7.3) | 28 (0.2) | 121 (0.9) | 107 (8.3) |
| Female | 9,531 (42.5) | 624 (6.5) | 504 (5.3) | 31 (0.3) | 89 (0.9) | 78 (8.2) |
| Histology type, n (%) | ||||||
| Non-small cell lung cancer | 8,732 (38.9) | 764 (8.7) | 613 (7.0) | 30 (0.3) | 42 (0.5) | 79 (0.9) |
| Small cell lung cancer | 165 (0.7) | 138 (83.6) | 113 (68.5) | 0 | 25 (15.2) | 0 |
| Colorectal cancer | 3,720 (16.6) | 61 (1.6) | 44 (1.2) | 0 | 1 (<0.1) | 16 (0.4) |
| Hepatocellular carcinoma | 1,649 (7.4) | 233 (14.1) | 167 (10.1) | 12 (0.7) | 38 (2.3) | 16 (1.0) |
| Gastric cancer | 1,431 (6.4) | 38 (2.7) | 25 (1.7) | 1 (0.1) | 1 (0.1) | 11 (0.8) |
| Biliary tract cancer | 1,369 (6.1) | 69 (5.0) | 50 (3.7) | 3 (0.2) | 4 (0.3) | 12 (0.9) |
| Pancreatic cancer | 910 (4.1) | 32 (3.5) | 18 (2.0) | 1 (0.1) | 1 (0.1) | 12 (1.3) |
| Breast cancer | 661 (2.9) | 62(9.4) | 36 (5.4) | 5 (0.8) | 14 (2.1) | 7 (1.1) |
| Ovarian cancer | 561 (2.5) | 31 (5.5) | 16 (2.9) | 2 (0.4) | 7 (1.2) | 6 (1.1) |
| Sarcomas | 554 (2.5) | 71 (12.8) | 33 (6.0) | 3 (0.5) | 31 (5.6) | 4 (0.7) |
| Kidney cancer | 545 (2.4) | 13 (2.4) | 8 (1.5) | 0 (0.0) | 1 (0.2) | 4 (0.7) |
| Prostate cancer | 356 (1.6) | 22 (6.2) | 8 (2.2) | 0 (0.0) | 9 (2.5) | 5 (1.4) |
| Esophageal cancer | 293 (1.3) | 32 (10.9) | 21 (7.2) | 2 (0.7) | 8 (2.7) | 1 (0.3) |
| Cervical cancer | 275 (1.2) | 18 (6.5) | 14 (5.1) | 0 (0.0) | 2 (0.7) | 2 (0.7) |
| Endometrium cancer | 252 (1.1) | 21 (8.3) | 15 (6.0) | 0 (0.0) | 4 (1.6) | 2 (0.8) |
| GIST | 236 (1.1) | 19 (8.1) | 14 (5.9) | 0 (0.0) | 3 (1.3) | 2 (0.8) |
| Melanoma | 235 (1.0) | 6 (2.6) | 5 (2.1) | 0 (0.0) | 0 (0.0) | 1 (0.4) |
| Bladder cancer | 209 (0.9) | 40 (19.1) | 24 (11.5) | 0 (0.0) | 12 (5.7) | 4 (1.9) |
| Neuroendocrine cancer | 170 (0.8) | 40 (23.5) | 32 (18.8) | 0 (0.0) | 7 (4.1) | 1 (0.6) |
| Thyroid cancer | 87 (0.4) | 1 (1.1) | 1 (1.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Head and neck cancer | 22 (0.1) | 1 (4.5) | 1 (4.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Classification of RB1 mutation according to ACMG guidelines, n (%) | ||||||
| P | 1,204 (70.3) | 966 (56.4) | 25 (1.46) | 210 (12.3) | 3 (0.1) | |
| LP | 66 | 33 (1.9) | 12 (0.7) | 15 (0.9) | 0 (0.0) | 6 (0.4) |
| VUS | 461 (26.9) | 271 (15.8) | 19 (1.1) | 0 (0.0) | 171 (10.0) | |
| LB | 3 (0.2) | 3 (0.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| B | 7 (0.4) | 2 (0.1) | 0 (0.0) | 0 (0.0) | 5 (0.3) | |
| N/A | 4 (0.2) | 4 (0.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| MSI status, n (%) | ||||||
| MSI-H | 458 (3.0) | 31 | 31 (6.8) | 0 (0.0) | 0 (0.0) | |
| MSS | 14,626 (65.0) | 754 (5.2) | 36 (0.2) | 171 (1.2) | ||
| N/A | 7,348 (33.0) | 473 (6.4) | 23 (0.3) | 39 (0.5) | ||
ACMG, American College of Medical Genetics; B, benign, GIST, gastrointestinal stromal tumor; LB, likely benign; LP, likely pathogenic; MSI-H, high microsatellite instability; MSS, microsatellite stability; N/A, not applicable; P, pathogenic; SNV, single-nucleotide variant; VUS, variants of uncertain significance.
We have compared difference between our data and the data of TCGA, of which 10 cancer types are included. In the TCGA database, RB1 mutations occurred in 15.5%, 5.3%, and 3.5% of endometrial, melanoma, and colorectal cancers, respectively, which were numerically higher than in our cohort. The RB1 frequencies in the remaining cancer types were similar between TCGA and our datasets. SNVs were the predominant variant types for RB1 in both datasets (Table 2).
Table 2
| Histology type, n (%) | No. (Chinese/TCGA) |
RB1 mutation | RB1 SNV | RB1 loss | |||||
|---|---|---|---|---|---|---|---|---|---|
| Chinese | TCGA | Chinese | TCGA | Chinese | TCGA | ||||
| Non-small cell lung cancer | 8,732/1,089 | 764 (8.7) | 60 (5.5) | 613 (7.0) | 41 (3.8) | 42 (0.5) | 17 (1.6) | ||
| Colorectal cancer | 3,720/633 | 61 (1.6) | 22 (3.5) | 44 (1.2) | 19 (3.0) | 1 (<0.1) | 3 (0.5) | ||
| Gastric cancer | 1,431/433 | 38 (2.7) | 15 (3.5) | 25 (1.7) | 12 (2.8) | 1 (0.1) | 3 (0.7) | ||
| Hepatocellular carcinoma | 1,649/377 | 233 (14.1) | 22 (5.8) | 167 (10.1) | 12 (3.2) | 38 (2.3) | 1 (0.3) | ||
| Breast cancer | 661/1,098 | 62 (9.4) | 28 (2.6) | 36 (5.4) | 21 (1.9) | 14 (2.1) | 11 (1.0) | ||
| Endometrium cancer | 252/560 | 21 (8.3) | 87 (15.5) | 15 (6.0) | 76 (13.6) | 4 (1.6) | 13 (2.3) | ||
| Sarcomas | 554/261 | 71 (12.8) | 27 (10.3) | 33 (6.0) | 15 (5.7) | 31 (5.6) | 8 (3.1) | ||
| Melanoma | 235/470 | 6 (2.6) | 25 (5.3) | 5 (2.1) | 20 (4.3) | 0 (0.0) | 8 (1.7) | ||
| Cervical cancer | 275/307 | 18 (6.5) | 21 (6.8) | 14 (5.1) | 16 (5.2) | 2 (0.7) | 6 (2.0) | ||
| Head and neck cancer | 22/528 | 1 (4.5) | 16 (3.0) | 1 (4.5) | 13 (2.5) | 0 (0.0) | 4 (0.8) | ||
SNV, single-nucleotide variant.
RB1 loss occurred most frequently in SCLC (25/165, 15.2%), bladder cancer (12/209, 5.7%), and sarcomas (31/554, 5.6%); RB1 SNV was most common in SCLC, and fusions most commonly occurred in breast cancer (Figure 1A). There were 185/1,712 (10.8%) patient samples with germline RB1 mutations. Stratifying by the type of mutation, it was found that 1,443 (6.40%) patients had tumors with SNVs, 59 (0.26%) with fusions, and 210 (0.94%) with loss of RB1 (Figure 1B).
Of the 22,432 patients, microsatellite status was determined in 15,084 (67.2%); MSI-H accounted for 3% (458/15,084) and MSS for 65% (14,626/22,432). No tumors with an MSI-H status were identified in patients with RB1 fusion or RB1 loss, and MSI-H was found only in RB1 SNV tumors, suggesting that MSI-H might be correlated with the classification of RB1 mutation (Table 1).
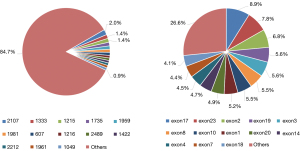
The clinical impact of the individual variants was divided into the following 5 types, according to the American College of Medical Genetics and Genomics guidelines: benign (B), likely benign (LB), variants of uncertain significance (VUS), likely pathogenic (LP), and pathogenic (P). In the present study, the P category accounted for the highest proportion of mutations (1,204/1,712, 70.3%) (Table 1). The pie chart (Figure 2) shows that the pathogenic mutations were quite homogeneously distributed along the gene at some somatic mutation hotspots (average hotspots accounted for 1% of all RB1 mutations. The site with the highest pathogenic mutation rate were 2,107 and 1,333, accounting for 2% and 1.4%, respectively (Figure 2A). For pathogenic exonic mutations, exons 17, 23, 2, 19, and 3 accounted for 8.9%, 7.8%, 6.8%, 5.6%, and 5.6%, respectively, being the top 5 with the highest mutation frequency (Figure 2B).
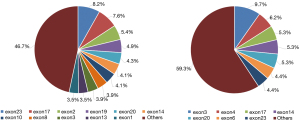
The cancers with the highest proportion of RB1 SNVs were SCLC and lung adenocarcinoma (LUAD), accounting for 68.5% and 36%, respectively (data not shown). Exons 23 and 17 of the RB1 gene were had the highest pathogenic mutation frequency in LUAD, accounting for 8.2% and 7.6%, respectively (Figure 3A). However, in SCLC, exon 3 and 4 mutations accounted for 9.7% and 6.2%, respectively (Figure 3B). Therefore, the location of the RB1 mutation could be associated with different lung cancer histologic classifications.
Discussion
To the best of our knowledge, the present study is the first study of wide-ranging RB1 aberrations in Chinese patients with solid tumors, determining the occurrence of RB1 mutations in many different solid cancer histotypes and their prevalence in China. We found widely distributed RB1 alterations in solid cancers among Chinese people, significantly differing among different tumor types, which provides further clinical evidence of RB1 mutation in cancer patients.
RB1 alterations were detected in 7.6% of our Chinese cancer patient cohort, a similar frequency to that previously reported worldwide (7.2% in 190,247 different solid tumors) (13). In our cohort, 83.6% of SCLC patients harbored RB1 mutations, a frequency much higher than in a recent study that reported a rate of 75% (14). Neuroendocrine cancers exhibited RB1 mutations in 23.5% of our cases, consistent with a previous study of 320 neuroendocrine neoplasm patients, in which the RB1 mutation rate was 19.7%, as detected by ctDNA NGS (15). However, evidence for RB1 mutations in Chinese population is still inadequate.
It is well known that RB1 mutations are present in different types of cancer, including SCLC, which makes it a potential therapeutic target. However, no drug can directly reactivate the RB1 gene and restore its normal function. Chemical and genetic fragility screens were performed by Lyu et al. who discovered that inhibiting Aurora kinase A (AURKA) resulted in synthetically lethal RB1 loss in lung cancer. Inhibiting AURKA reduced phosphorylation and activated the function of stathmin, resulting in an extremely unstable microtubule environment in RB1–/– cells, seriously impeding bipolar spindle formation and inducing cell death (16). In 2019, Owonikoko et al. conducted a prospective clinical trial to assess the efficacy of paclitaxel, together with the AURKA inhibitor alisertib, in 178 SCLC patients compared with paclitaxel and placebo, yielding a progression-free survival of 3.32 and 2.17 months, respectively, although there was no significant difference (P=0.113) (17). Furthermore, the benefit gap between the 2 treatment regimens increased and reached significance in patients with cell-cycle regulator mutations [including RB1, retinoblastoma-like (RBL)1 gene, RBL2, and CDK6) [alisertib + paclitaxel vs. paclitaxel and placebo, median progression-free survival (mPFS) 3.68 vs. 1.80 months, P=0.0003] (17). The results demonstrated that alisertib+paclitaxel seems to be effective as a second-line therapy for SCLC with RB1 mutations. For RB1-deficient SCLC, chemotherapy was initially effective, but the tumor rapidly became chemotherapeutic resistance, especially in SCLC patients with systematic treatment (18), which could be one of the most likely reasons for the poor efficacy of SCLC treatment. Gong et al. analyzed the LY3295668 AURKA inhibitor and found that it demonstrated strong inhibitory activity on tumor cell lines with RB1 mutations (19). The inhibitory action of LY3295668 on human bone marrow cells was 10-fold less than on tumor cell lines, indicating its potentially safe application (19). These studies suggest that the AURKA protein could be considered a therapeutic target in RB1-mutated cancer. The main mechanism of CDK4/6 inhibitors is to inhibit the phosphorylation of pRB and induce G1 cell-cycle blocking in tumor cells (20). In 2017, Goel demonstrated that selective CDK4/6 inhibitors could induce cell-cycle arrest, as well as mediate anticancer and stimulate the immune system in mouse models of breast cancer and other solid tumors (21). This finding suggests that CDK4/6 inhibitors combined with immunotherapy might be a promising treatment option for RB1-deficient tumors. Treatment with topoisomerase (TOP) inhibitors (irinotecan and indimitecan) on patient-derived xenografts (PDXs) of TNBC, results revealed that RB1 loss predict response to these TOP inhibitors (22). Although these data are currently available, but deeper insights for RB1 in cancer development still keeps being unclear. Besides, epidemiological data for RB1 mutation in cancer patients are also waiting to be further supplemented and improved.
CDK4/6 inhibition has been considered one of the most promising anticancer therapeutic strategies. In particular, most human cancer cell-cycle regulation pathway entry regulators are defective in mitosis initiators; for example, the D-type cyclins overexpression of, CDK4/6 amplification/mutation, dysfunction of cyclin that inhibit CDKN2 members, finally contributing to phosphorylating RB1 and leading to the unrestricted proliferation of tumor cells (23). Loss of RB1 function is one of the most important causes of acquired resistance to CDK4/6 inhibitors. When CDK4/6 are activated by D-type, these complexes of the RB1 family decrease the expression of cell cycle genes E-type cyclins inactivate RB1 by activating downstream kinases such as CDK2 and CDK1, which lead to DNA replication (S phase) and chromosome segregation (4). Thus, the RB1 signaling pathway is downstream of the CDK4/6 signaling pathways. Targeting the inhibition of the specific CDK/cyclin complex is designed as a potential intervention to regain RB1’s normal tumor suppressor function via blocking its phosphorylation (24). By analyzing more than 340 cancer-associated genes (MSK-IMPACT, MSKCC) in ER+ breast cancers, Li et al. identified RB1 alterations in addition to loss-of-function mutations in FAT1 as the most significant mechanism of resistance to CDK4/6 inhibitors (5). A recent study suggests that biallelic disruption of RB1 that enriched in the resistant tumors contributes to resistance of CDK4/6 inhibitor, highlighting the importance of testing RB1 status (6). Condorelli and his colleagues investigated breast cancer patients who had pre- and post-treatment genotyping in tissue and peripheral blood samples during treatment with CDK 4/6 inhibitors. In three patients exposed to CDK4/6 inhibitors, detectable acquired RB1 mutations were found in circulating tumor DNA (ctDNA) (7). In the O’Leary s cohort, RB1 mutations emerged only in the palbociclib plus fulvestrant arm and in a minority of metastatic breast cancer patients (8). Most studies have shown that CDK4/6 inhibitors are effective in RB1-intactive solid tumors (25). Palbociclib, ribociclib and abemaciclib are known CDK4/6 inhibitors, and all dephosphorylate RB1, which in turn activates the RB1 pathway to suppress tumors that have lost RB1 function. Recently, Gong et al. conducted a study to identify tumors that are more likely to be susceptible to CDK4/6 inhibitors, for this purpose, they used 560 cancer cells (26). Nearly all tumors were found to be sensitive to abemaciclib and expressed RB1 (27), although there was insufficient evidence to conclude that a high expression of RB1 was related to sensitivity to this drug. Rubio et al. confirmed that the state of RB1 was irrelevant for the sensitivity of bladder cancer cell lines to CDK4/6 inhibitors (28). This finding suggests that CDK4/6 inhibitors can be used to treat bladder cancer previously not suitable for current treatment regimens. A recent study showed that 3 patients with estrogen receptor (ER)+/human epidermal growth factor receptor 2 (HER-2) and new RB1 mutations in breast cancer caused failure of treatment with CDK4/6 inhibitors (palbociclib or ribociclib), suggesting that resistance to CDK4/6 inhibitors may be caused by RB1 mutations identified (7). Due to the low mutation frequency of RB1 after treatment, inactivation of RB1 is not the main cause of resistance to CDK4/6 inhibitors. Walter et al. showed that CDK2 was involved in the phosphorylation of RB1, which has been shown to result in resistance to CDK4/6 inhibitors in a murine model of KRAS mutated lung cancer (29). This suggests that simultaneously targeting CDK4/6 and CDK2 proteins can effectively reactivate the RB1 pathway. CDK4/6 inhibitor resistance associated with RB1 mutations needs further clinical research to explore treatment modalities. In the present study, we also observed widely distributed RB1 alterations in solid tumors. However, we did not investigate whether RB1 mutations were associated with CDK4/6 inhibitor resistance, which warrants further research.
The functional integrity of RB1 is important for the effectiveness of immunotherapy. Furthermore, it has recently been suggested that RB1 mutations in patients with non-SCLC are associated with unresponsiveness to nivolumab and pembrolizumab immunotherapy (14). In the study by Bhateja et al., of 66 patients receiving immunotherapy, 6 with RB1 mutations did not respond to treatment (14). In addition, RB1 can affect the changes of tumor microenvironment caused by CDK4/6 inhibitors and DNA-damaging agents, especially the reduction of programmed cell death-ligand 1 expression by phosphorylating RB1 which has an impact on the treatment efficacy of CDK4/6 inhibitors and DNA-damaging agents (30). Göran Jönsson et al. revealed 23 deleted regions, many of which have been previously shown to be characteristics of BRCA1-mutant breast cancer, including the RB1 region of 13q14.2 (31). RB1 mutations were more frequent in tumors with alterations in BRCA1 (12/33, 36%) than in BRCA2-mutated tumors (1/33, 3%), and physical disruption of RB1 was observed in BRCA1-deficient cell lines (32). These results suggest that loss of chromosome 13 plays a vital role in the development of breast cancer in patients with BRCA1 mutation, potentially by inactivating RB1.
Next-generation sequencing (NGS) has high sensitivity and specificity. Previous studies have compared the performance of three different techniques of testing gene mutations. NGS, immunohistochemistry (IHC), and fluorescence in situ hybridization (FISH) share more than 98% identity for detecting copy number variation (33). Hovelson and his colleagues developed the Oncomine Comprehensive Panel (OCP), an integrative NGS-based assay, which achieved >95% accuracy for KRAS, EGFR, and BRAF mutation detection (34). For the NGS panel employed in our study, the maximal sensitivities for detecting single nucleic variances (SNVs) and small insertions/deletions (Indels) reached 99% and 98.7%, respectively. For SNV and indel detection in clinical samples, targeted NGS can identify all hotspot mutations with 100% sensitivity and specificity (11). The results showed a concordance rate of 100% compared to the amplification refractory mutation system (ARMS) (11). Therefore, we chose NGS, which has high sensitivity and specificity, to detect RB1 mutations based on its good performance.
The present study has several limitations. First, the cancer types were not characterized in any detail, which could lead to possible bias. Second, there were no survival data for the cross-sectional setting of the study, limiting any conclusions on the possible association between changes in RB1 and prognosis or response of treatment. Third, we did not investigate whether RB1 mutations were associated with CDK4/6 inhibitor resistance. These warrant further investigation.
Conclusions
The findings of the present study indicate that alterations of RB1 occur in different cancer types in Chinese solid tumor patients, and frequencies of specific alterations differ markedly among tumor types. Our research suggests that NGS should be used when a personalized characterization of cancer is required and after molecular profiling is performed in a clinical treatment environment. Further research on personalized cancer treatment is warranted.
Acknowledgments
Funding: This work was supported by a grant from the National Natural Science Foundation of China (No. 82172973), the Key Research and Development Project of Shaanxi Province (No. 2022ZDLSF03-04), and the Key Clinical High Tech Project of Xijing Hospital (No. XJZT19Z26).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3162/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3162/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3162/coif). XS, WD, HZ, DH, TB, and DF report that they are the employees of The Medical Department, 3D Medicines Inc. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics committee of the First Affiliated Hospital of the Air Force Medical University (No. KY20222189-F-1), and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Flores M, Goodrich DW. Retinoblastoma Protein Paralogs and Tumor Suppression. Front Genet 2022;13:818719. [Crossref] [PubMed]
- Valverde JR, Alonso J, Palacios I, et al. RB1 gene mutation up-date, a meta-analysis based on 932 reported mutations available in a searchable database. BMC Genet 2005;6:53. [Crossref] [PubMed]
- Dyson NJ. RB1: a prototype tumor suppressor and an enigma. Genes Dev 2016;30:1492-502. [Crossref] [PubMed]
- Álvarez-Fernández M, Malumbres M. Mechanisms of Sensitivity and Resistance to CDK4/6 Inhibition. Cancer Cell 2020;37:514-29. [Crossref] [PubMed]
- Li Z, Razavi P, Li Q, et al. Loss of the FAT1 Tumor Suppressor Promotes Resistance to CDK4/6 Inhibitors via the Hippo Pathway. Cancer Cell 2018;34:893-905.e8. [Crossref] [PubMed]
- Wander SA, Cohen O, Gong X, et al. The Genomic Landscape of Intrinsic and Acquired Resistance to Cyclin-Dependent Kinase 4/6 Inhibitors in Patients with Hormone Receptor-Positive Metastatic Breast Cancer. Cancer Discov 2020;10:1174-93. [Crossref] [PubMed]
- Condorelli R, Spring L, O'Shaughnessy J, et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann Oncol 2018;29:640-5. [Crossref] [PubMed]
- O'Leary B, Cutts RJ, Liu Y, et al. The Genetic Landscape and Clonal Evolution of Breast Cancer Resistance to Palbociclib plus Fulvestrant in the PALOMA-3 Trial. Cancer Discov 2018;8:1390-403. [Crossref] [PubMed]
- Egger JV, Lane MV, Antonucci LA, et al. Dephosphorylation of the Retinoblastoma protein (Rb) inhibits cancer cell EMT via Zeb. Cancer Biol Ther 2016;17:1197-205. [Crossref] [PubMed]
- Knudsen ES, Pruitt SC, Hershberger PA, et al. Cell Cycle and Beyond: Exploiting New RB1 Controlled Mechanisms for Cancer Therapy. Trends Cancer 2019;5:308-24. [Crossref] [PubMed]
- Su D, Zhang D, Chen K, et al. High performance of targeted next generation sequencing on variance detection in clinical tumor specimens in comparison with current conventional methods. J Exp Clin Cancer Res 2017;36:121. [Crossref] [PubMed]
- Zhou S, Zhang D, Li J, et al. Landscape of RAS Variations in 17,993 Pan-cancer Patients Identified by Next-generation Sequencing. Pathol Oncol Res 2020;26:2835-7. [Crossref] [PubMed]
- Jardim DL, Millis SZ, Ross JS, et al. Cyclin Pathway Genomic Alterations Across 190,247 Solid Tumors: Leveraging Large-Scale Data to Inform Therapeutic Directions. Oncologist 2021;26:e78-89. [Crossref] [PubMed]
- Bhateja P, Chiu M, Wildey G, et al. Retinoblastoma mutation predicts poor outcomes in advanced non small cell lung cancer. Cancer Med 2019;8:1459-66. [Crossref] [PubMed]
- Zakka K, Nagy R, Drusbosky L, et al. Blood-based next-generation sequencing analysis of neuroendocrine neoplasms. Oncotarget 2020;11:1749-57. [Crossref] [PubMed]
- Lyu J, Yang EJ, Zhang B, et al. Synthetic lethality of RB1 and aurora A is driven by stathmin-mediated disruption of microtubule dynamics. Nat Commun 2020;11:5105. [Crossref] [PubMed]
- Owonikoko TK, Niu H, Nackaerts K, et al. Randomized Phase II Study of Paclitaxel plus Alisertib versus Paclitaxel plus Placebo as Second-Line Therapy for SCLC: Primary and Correlative Biomarker Analyses. J Thorac Oncol 2020;15:274-87. [Crossref] [PubMed]
- Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer 2017;17:725-37. [Crossref] [PubMed]
- Gong X, Du J, Parsons SH, et al. Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of the RB1 Tumor Suppressor Gene. Cancer Discov 2019;9:248-63. [Crossref] [PubMed]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 1999;13:1501-12. [Crossref] [PubMed]
- Goel S, DeCristo MJ, Watt AC, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 2017;548:471-5. [Crossref] [PubMed]
- Coussy F, El-Botty R, Château-Joubert S, et al. BRCAness, SLFN11, and RB1 loss predict response to topoisomerase I inhibitors in triple-negative breast cancers. Sci Transl Med 2020;12:eaax2625. [Crossref] [PubMed]
- Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov 2009;8:547-66. [Crossref] [PubMed]
- Indovina P, Pentimalli F, Casini N, et al. RB1 dual role in proliferation and apoptosis: cell fate control and implications for cancer therapy. Oncotarget 2015;6:17873-90. [Crossref] [PubMed]
- Knudsen ES, Witkiewicz AK. The Strange Case of CDK4/6 Inhibitors: Mechanisms, Resistance, and Combination Strategies. Trends Cancer 2017;3:39-55. [Crossref] [PubMed]
- Gong X, Litchfield LM, Webster Y, et al. Genomic Aberrations that Activate D-type Cyclins Are Associated with Enhanced Sensitivity to the CDK4 and CDK6 Inhibitor Abemaciclib. Cancer Cell 2017;32:761-776.e6. [Crossref] [PubMed]
- Klein ME, Kovatcheva M, Davis LE, et al. CDK4/6 Inhibitors: The Mechanism of Action May Not Be as Simple as Once Thought. Cancer Cell 2018;34:9-20. [Crossref] [PubMed]
- Rubio C, Martínez-Fernández M, Segovia C, et al. CDK4/6 Inhibitor as a Novel Therapeutic Approach for Advanced Bladder Cancer Independently of RB1 Status. Clin Cancer Res 2019;25:390-402. [Crossref] [PubMed]
- Walter DM, Yates TJ, Ruiz-Torres M, et al. RB constrains lineage fidelity and multiple stages of tumour progression and metastasis. Nature 2019;569:423-7. [Crossref] [PubMed]
- Jin X, Ding D, Yan Y, et al. Phosphorylated RB Promotes Cancer Immunity by Inhibiting NF-κB Activation and PD-L1 Expression. Mol Cell 2019;73:22-35.e6. [Crossref] [PubMed]
- Jönsson G, Staaf J, Vallon-Christersson J, et al. Genomic subtypes of breast cancer identified by array-comparative genomic hybridization display distinct molecular and clinical characteristics. Breast Cancer Res 2010;12:R42. [Crossref] [PubMed]
- Jönsson G, Staaf J, Vallon-Christersson J, et al. The retinoblastoma gene undergoes rearrangements in BRCA1-deficient basal-like breast cancer. Cancer Res 2012;72:4028-36. [Crossref] [PubMed]
- Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023-31. [Crossref] [PubMed]
- Hovelson DH, McDaniel AS, Cani AK, et al. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia 2015;17:385-99. [Crossref] [PubMed]


