
Role of gga-miR-29b-3p in suppressing the proliferation, invasion and migration of MSB1 Marek’s disease tumor cells by the targeting of the DNMT3B gene
Introduction
Marek’s disease (MD) is infectious disease in poultry that is caused by a highly cell-bound herpesvirus [MD virus (MDV)], which was characterized by immunosuppression and polyneuritis (1,2). It has been found across a range of commercial poultry species, in chickens and quail initially and later in turkeys (3,4). Live attenuated vaccination is still used worldwide at this stage in the prevention of MD (5). However, the widespread use of the vaccine has also led to the increased virulence of MDV, from a strong strain (vMDV) in the 1970s that broke through the contemporaneous turkey herpesvirus (HVT) vaccine, to a super-virulent strain (vvMDV) in the 1980s, and then to an very virulent plus (vv+MDV) in the 1990s that also broke through the protection of vaccines such as bivalent vaccines HVT+MDV2 (SB1) (6). The emergence of extra-virulent viruses and the increased virulence of wild strains have emerged as the major main challenges to the sustained control of this disease, causing considerable harm to livestock and economic losses in the poultry farming industry (7,8).
MicroRNAs (miRNAs), are short-stranded RNAs that are found endogenously in eukaryotes, consist of highly conserved sequences, and participate in the regulation of numerous biological processes (9). MiRNAs contribute to gene transcriptional regulation by degrading messenger RNA (mRNAs), inhibiting protein synthesis and interacting with long non-coding RNA (10,11). Recent reports have revealed that aberrant miRNA expression in MD may be involved in the tumorigenesis process. For instance, Gallus gallus (gga)-miR-21 was found to be upregulated during gallid herpesvirus 2 (GaHV-2) infection driven by activator protein 1 (AP-1) and Ets-response elements and to promote tumor cell growth and apoptotic escape by targeting the programmed death cell 4 gene (12). Another study reported that gga-miR-26a was significantly downregulated in the spleen of MDV-infected chickens and that it inhibited MSB1 cell proliferation in a direct targeting relationship with NEK6, possibly by suppressing prematurely senescent cells to affect the proliferative potential of tumor cells (13). Other reports revealed that reduced expression of gga-miR-103-3p in MDV-infected tissues and this miRNA inhibited MSB1 migration but had no significant effect on proliferation, with the abnormal expression of 2 target genes, CCNE1 and TFDP2, being shown to disrupt the normal cell cycle and thus precipitating tumorigenesis (14). Downregulation of gga-miR-130b-3p in MD tumor may be directly attributed to the methylation of this miRNA (15). Indeed, gga-miR-155 is considered to be a potential marker for MD tumor, and studies suggest that the overexpression of this miRNA has a beneficial influence on MSB1 cell proliferation, migration, and invasion, but has a suppressive effect on cell apoptosis by targeting RORA (16,17). Our previous miRNAs profile found that gga-miR-29b-3p was significantly downregulated in MD tumorous tissues, implicating it may be involved in MD tumorigenesis (18). In chicken primordial germ cells, gga-miR-29b was found to downregulate the expression of DNMT3B, which provides more knowledge to explain the role of DNMT genes regulation in embryogenesis (19). In the jejunal mucosa of laying hens with different genetic backgrounds, the potential target genes of miR-29b-3p were enriched in energy pathways at different stages of production periods (20). In the present study, we examined a mechanism of gga-miR-29b-3p in MD tumor transformation and its effects on the characteristics of a MDV-transformed lymphoid cell line. It is hoped our findings can contribute to building a theoretical foundation and identifying molecular markers for the designed breeding of MD disease resistance. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3519/rc).
Methods
Cell culture, miRNA transfection, and RNA interference
The MDV-transformed chicken lymphoblastoid cell line MDCC-MSB1 was kindly provided by Dr. C. Itakura in Tottori University and cultured at 37 ℃, under 5% CO2 in RPMI 1640 medium (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen) (21,22). X-tremeGENE siRNA Transfection Reagent (Roche, Basel, Switzerland) was selected to transfect the corresponding miRNA or siRNA into MSB1. The gga-miR-29b-3p mimics, inhibitor, DNMT3B small interfering RNA (siRNA), and scramble nontarget negative control (NC) were synthesized by GenePharma Company (GenePharma Co. Ltd., Shanghai, China). The targeting sequence of DNMT3B was 5'-GCTGTGCCTTGAACATTGT-3'. As described previously, dosage of mimics, siRNA, and corresponding NC transfection was 100 nM, while that of the inhibitor and corresponding NC transfection was 150 nM (13,23).
Detection of tumorous cellular functions
The cells used for transfection were seeded into 96-well, 24-well, and 6-well plates with a total of 3×104, 1.5×105, and 5×105 cells per well, respectively. Cell proliferation was detected using Cell Counting Kit-8 (CCK-8; Beyotime, Shanghai, China). The absorbance at 450 nm was examined at 24, 36, 48, 60, and 72 hours posttransfection. Cell apoptosis was detected using the TUNEL BrightRed Apoptosis Detection Kit (Vazyme, Nanjing, China). The cells were collected for apoptosis assay 48 hours posttransfection according to the manufacturer’s instructions (24). Briefly, the cells were fixed with 4% paraformaldehyde at 4 ℃ for 20 minutes and then permeabilized with 0.5% Triton X-100 at room temperature for 5 minutes. The cells were next labeled with tetramethylrhodamine (TMR) red deoxyuridine triphosphate (dUTP) which could bind with the broken DNA in the apoptotic cells, and the reaction was terminated with 20 mM of ethylenediaminetetraacetic acid (EDTA). Following this, the cells were stained with 1 µg/mL of DAPI. Finally, the cells were observed by fluorescence microscope (Olympus Corporation, Tokyo, Japan) and photographed. The procedures of the cell cycle assay were performed according the specifications of Li et al. [2017] and Zhao et al. [2017] (23,25). After fixation, the cells were treated with RNase A (20 µg/mL; Sigma-Aldrich, St. Louis, MO, USA) and then stained with propidium iodide (50 µg/mL; Sigma-Aldrich) for 30 minutes in the dark. The cell populations were quantified by FlowJo software (BD, Franklin Lakes, NJ, USA). The protocols of cell migration assay were also performed according to those of Li et al. [2014] (13).
RNA extraction and real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol Reagent (Invitrogen), and complement DNA (cDNA) was synthesized using a PrimeScript RT Reagent Kit (Takara Bio, San Jose, CA, USA). The RT-qPCR reactions were performed using the SYBR Premix Ex Taq II kit (Takara Bio) and detected on the ABI Prism 7500 HT sequence detection system (Applied Biosystems, Foster City, CA, USA). The primer information of matrix metallopeptidase (MMP) MMP2, MMP9, and β-actin can be found in the publication of Zhao et al. [2017] (Table S1) (23); that for MDV Eco Q fragment-encoded gene MEQ from Heidari et al. [2008] (26); that for BCL2 and BCL2L1 from Subramaniam et al. [2013] (27); that for TNFSF10 from Li et al. [2008] (28); and that for the DNMT3B gene from Tian et al. [2013] (Table S1) (29). The results were determined by the 2–ΔΔCt method (30).
Western blot
Western blot was conducted using standard methods (31) with specific primary antibody to DNMT3B (1:2,000; Abbkine, Wuhan, China) or β-actin antibody (1:1,000; Beyotime) overnight, along with anti-rabbit secondary antibody (1:1,000; Beyotime) and anti-mouse secondary antibody (1:1,000; Beyotime). The specific protein was visualized with SuperSignal West Pico PLUS (Life Technologies, Thermo Fisher Scientific). Grayscale analysis was performed using Image J software (National Institutes of Health, Bethesda, MD, USA).
Target gene prediction and dual-luciferase reporter assay
Potential miRNA targets of gga-miR-29b-3p were predicted the by online software platforms TargetScan (http://www.targetscan.org/) and miRDB (http://mirdb.org/miRDB/).
The 3 prime untranslated region (3'-UTR) fragments of DNMT3B covering the putative gga-miR-29b-3p binding sites were amplified to construct the wild-type and mutant-type luciferase reporter vectors. This region was amplified using forward primer 5'-GCCTCGAGCGTTTCAAAATGCTGCTG-3' and reverse primer 5'-CGGCGGCCGCTTTTTTTTTTCCTCTTATTTTAAAAGATTGTATA-3'. The mutant-type vector was also constructed to further verify the binding sites between gga-miR-29b-3p and the 3'-UTR of DNMT3B. The sequence of seed region in the 3'- UTR of DNMT3B was completely mutated with forward primer 5'-TCTAACTCTCCCACCACGATTTTTTTTAGTTAAAGGAT-3' and reverse primer 5'-TAACTAAAAAAAATCGTGGTGGGAGAGTTAGAAACCTT-3'. The amplified fragments were inserted into the psiCHECK2 plasmid.
The dual-luciferase reporter experiments were performed in the 293T cell line as described previously (14). The 293T cell line was cultured in DMEM medium (Invitrogen) with 10% FBS (Invitrogen), which was maintained at the condition of 37 ℃ and 5% CO2. The luciferase activity was measured using the dual-luciferase Reporter Assay System (Promega, Madison, WI, USA).
Statistical analysis
Numerical data are presented as mean ± standard error (SE). All statistical analyses were conducted with the SPSS 22.0 software (IBM Corp., Armonk, NY, USA). The Student’s t-test and ANOVA test were performed for data of two groups and over two groups., respectively. A P value <0.05 was considered to indicate a significant difference, while a P value <0.01 was considered to indicate an extremely significant difference.
Results
gga-miR-29b-3p inhibited MSB1 proliferation by promoting apoptosis-independent cell cycle retardation
To understand the role of gga-miR-29b-3p in the MSB1 cell line, we used gga-miR-29b-3p mimics or inhibitor to overexpress or suppress miRNA expression in MSB1 cells, respectively. First, high transfection efficiency was verified (Figure 1A). Transfection of gga-miR-29b-3p mimics significantly inhibited MSB1 cell proliferation compared with NC mimics at 24, 36, 48, and 72 hours. In contrast, the transfection of gga-miR-29b-3p inhibitor significantly increased MSB1 cell proliferation compared with NC inhibitor at 48 and 60 hours (Figure 1B).

To further analyze whether the gga-miR-29b-3p-impaired cell proliferation was caused by apoptosis in MSB1 cells, we performed fluorescent staining and analyzed the gene expression level of endogenous and exogenous apoptotic pathways. Using staining by cellular fluorescence, we found that the rates of apoptotic cells increased significantly after transfection with miRNA mimics and decreased significantly after transfection with miRNA inhibitor (Figure 1C,1D). BCL2, BCL2L1, and TNFSF10 are key genes in the apoptotic pathway and necessary to induce cell apoptosis. We detected the expression levels of these genes. At 24 hours, the transcriptional expression of antiapoptotic BCL2 was significantly downregulated and that of antiapoptotic BCL2L1 was extremely significantly decreased; at 72 hours, after overexpression of gga-miR-29b-3p mimics, both the transcriptional expressions of BCL2 and BCL2L1 were significantly decreased (Figure 1E,1F). Under the condition of transfection with gga-miR-29b-3p inhibitor, BCL2 was significantly increased at 24 hours and significantly increased at 72 hours, while BCL2L1 was significantly increased at 24 hours (Figure 1E,1F). The expression of the proapoptotic gene TNFSF10 was significantly increased by transfection of gga-miR-29b-3p mimics at 24 hours and was extremely decreased by transfection of gga-miR-29b-3p inhibitor at 48 hours (Figure 1G). Moreover, there was no significant effect on cell cycle at 48 hours after gga-miR-29b-3p mimic or inhibitor transfection (Figure 1H,1I). This suggested that gga-miR-29b-3p played a major role in promoting apoptosis through regulating the key gene expression level in the apoptotic pathway in MSB1 cells.
gga-miR-29b-3p inhibited the expression of invasion-related genes and MSB1 cell migration
Migration and invasion are among the malignant behaviors of tumor cells. Invasion and migration have different biological characteristics: the former involves the occupation of malignant tumors, both primary or secondary, to adjacent host tissues, while the latter involves the transfer of tumor cells to another site or organ by blood flow and the continued growth of tumors. To further clarify the roles of gga-miR-29b-3p, we selected the invasion markers MMP2 and MMP9 for comparing the ability of invasion between the transfection of gga-miR-29b-3p and controls. Interestingly, RT-qPCR showed the transcription level of MMP2 to be significantly decreased after transfection of gga-miR-29b-3p mimics at 48 hours, while the opposite phenomenon of significantly upregulated transcription levels in MMP2 was observed under transfection of gga-miR-29b-3p inhibitor at 24 and 48 hours (Figure 2A). Moreover, the transcription level of MMP9 was downregulated significantly at 72 hours, while the transcription level of MMP9 was significantly increased posttransfection with miRNA inhibitor at 48 and 72 hours (Figure 2B). The cell number of migrated MSB1 significantly decreased under transfection of gga-miR-29b-3p mimics and was rescued by gga-miR-29b-3p inhibitor (Figure 2C,2D). This indicated that gga-miR-29b-3p restrained MSB1 cell invasion and migration.

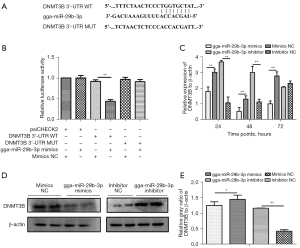
gga-miR-29b-3p suppressed DNMT3B gene expression upon binding to its 3'-UTR sequence
To elucidate effect of gga-miR-29b-3p in MD tumor formation, we next used online software and literature retrieval to select candidate gene DNMT3B. To reveal whether gga-miR-29b-3p could directly regulate its hypothetical candidate gene, DNMT3B, we constructed vectors including gga-miR-29b-3p wild binding site [wild type (WT)] and mutant binding site (MUT; Figure 3A). Luciferase activity showed that gga-miR-29b-3p had a distinctly inhibitory role on the WT group but not the MUT group (Figure 3B). To further validate that DNMT3B was directly regulated by gga-miR-29b-3p, we collected MSB1 cells under transfection with gga-miR-29b-3p mimics, inhibitor, and respective NCs at different time points to compare the expression of DNMT3B at the transcription level and protein level. First, the transcription level of DNMT3B was remarkably suppressed in the mimic transfection group at all time points of received samples but was extremely elevated at 24 and 48 hours in the inhibitor transfection group (Figure 3C). Second, the expression at translational levels of DNMT3B was remarkably inhibited in the mimic transfection group and extremely elevated in the inhibitor transfection group at 96 hours (Figure 3D,3E). Collectively, these results suggested that DNMT3B was directly prohibited by gga-miR-29b-3p and rescued through inhibitor.
DNMT3B knockdown inhibited cell proliferation by the promoting apoptosis of MSB1 cells
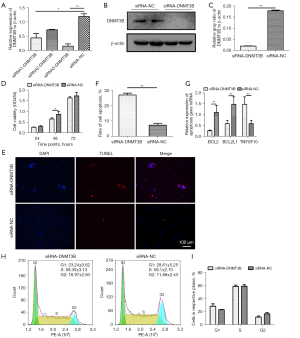
After identifying DNMT3B as a direct target gene of gga-miR-29b-3p, we analyzed its function in MD tumorigenesis by RNA interference. We designed 3 siRNA sequences to intervene with the DNMT3B gene, with the interference efficiency of the third sequence being almost 67% at the transcriptional level (Figure 4A). Using this RNA (hereafter namely siRNA-DNMT3B), the expression level of DNMT3B decreased by 87.5% at the translational level (Figure 4B,4C). Transfection of siRNA-DNMT3B at 48 hours significantly inhibited cell proliferation (Figure 4D). TUNEL assay showed that the number of apoptotic cells increased significantly after transfection with siRNA-DNMT3B (Figure 4E,4F). The DNMT3B gene intervention significantly downregulated the expression of the BCL2 and BCL2L1 genes but upregulated the mRNA expression of the TNFSF10 gene (Figure 4G). In contrast, knockdown of DNMT3B had no impact on cell cycle (Figure 4H,4I). Taken together, our findings revealed that silencing DNMT3B could stimulate spontaneous apoptosis and lead to cell cycle-independent proliferation in MSB1 cells.
Silencing of DNMT3B impaired the expression of invasion-related genes
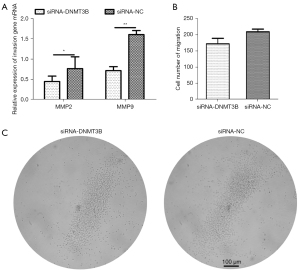
To further verify whether the gga-miR-29b-3p target gene, DNMT3B, suppresses invasion and migration, we chose 2 indicators, cell migration and invasion-related genes (MMP2 and MMP9), to examine in follow-up experiments of DNMT3B knockdown of MSB1 cells. First, the transcription level of MMP2 and MMP9 was visibly downregulated in DNMT3B knockdown at 48 hours (Figure 5A). Second, the cell numbers of migration were not obviously different after transfection with siRNA-DNMT3B at 64 hours (Figure 5B,5C). Therefore, we concluded that DNMT3B mediated invasion and exerted an enhanced effect on the viability MSB1 cells.
gga-miR-29b-3p and DNMT3B gene affected the expression of the proto-oncogene MEQ
Among the genes encoded by MDV, MEQ is considered to be an important oncogene. We observed that gga-miR-29b-3p and its target gene DNMT3B could affect MEQ expression. MEQ expression was significantly downregulated after transfection with gga-miR-29b-3p mimics at 48 hours. Conversely, MEQ expression was visibly increased after transfection with gga-miR-29b-3p inhibitor at 48 and 72 hours (Figure 6A). Moreover, MEQ expression significantly decreased after DNMT3B interference at 48 hours (Figure 6B). These observations indicated that gga-miR-29b-3p and its target gene, DNMT3B, inhibited the expression of genes involved in proto-oncogenesis.

Discussion
MD is a type of lymphoproliferative disease resulting from a cell-associated pathogen MDV and is capable of causing massive damage to the poultry industry (7). Once birds are infected with MDV, culling and mortality occur in up to 80% of the population, and there remains no viable cure for this disease anywhere in the world (32). Therefore, there is an urgent need to conduct further research to discover any potential molecular mechanisms of MD and to develop more promising therapeutic approaches. In our previous study using Solexa deep sequencing, we observed the aberrant expression of gga-miR-29b-3p in the MD tumorous spleen and liver lymphoma, and its expression pattern was negatively correlated with MD (18). We conducted further study on gga-miR-29b-3p-mediated effects to better understand the link between this miRNA and MD tumorigenesis.
MiRNAs have important functions in the proliferation, migration, invasion, and chemoresistance of various solid tumors (33). Recent studies have indicated that miR-29b shows aberrant expression in a variety of tumorous tissues and tumorous cells, which indicates that miR-29b may play a vital role during carcinogenesis (34,35). As per previous reports, miR-29b participates in the differentiation and tumorigenesis of tumor in prostate cancer, breast cancer, glioblastoma multiforme (GBM), and non-small cell lung cancer (NSCLC), implicating miR-29b as a central component in tumor progression (36-39). A study in miR-29b in nude mice revealed that the overexpression of miR-29b could limit the metastasis of prostate tumor cells and inhibit the growth of prostate xenografts, which was consistent with our findings related to gga-miR-29b-3p (36). In another study, miR-29b was shown to inhibit antiangiogenesis and antitumorigenesis by targeting AKT3 and inducing the expression of VEGF and C-myc in breast cancer cells (38). MiR-29b was demonstrated to affect the self-renewal and proliferation in neural stem cells (NSCs) by regulating the β-catenin and T-cell factor-mediated Wnt/β-catenin signaling pathway (40). The genetic developmental mechanism of NSC-transformed GBM was similar with NSCs; it was reported that miR-29b had the ability to inhibit cell growth, induce apoptosis and produce anticancer effects in GBM cell lines (39). Moreover, gga-miR-29b-3p was found to inhibit the proliferation of an MDV-derived chicken lymphocyte line, MDCC-MSB1, and the decrease in cell proliferation rate resulted from an elevated number of apoptotic cells; the apoptotic pathways included exogenous pathway triggered by death receptors and endogenous pathway triggered by mitochondria (41). The exogenous apoptotic pathway primarily consists of a combination of death receptors and ligands leading to the receptor multimerization, in which TNFSF10 acts as a ligand that binds to death receptors DR4 and DR5 and activates caspase-8 to trigger the apoptotic cascade reaction (42). The B-cell lymphoma 2 (BCL2) family regulates the permeability of the mitochondrial outer membrane by polymerizing and depolymerizing with the components in this family (43).
Based on our findings, we concluded that the overexpression of gga-miR-29b-3p suppressed BCL2 and BCL2L1 and promoted the expression of the proapoptotic gene, TNFSF10. Invasion and migration are 2 powerful functions of tumor cells, in which MMPs are involved (44). Among the MMP family, MMP2 and MMP9 perform the effect of degrading type IV collagen, a major component of the basement membrane, prompting tumorous cell invasion in cancer (45). In one study, overexpression of miR-29b in vitro seemed to strengthen NSCLC cell proliferation, migration, and invasion by elevating cyclin D1 or MMP9 and reducing the expression of p21, Bax, and E-cadherin; moreover, the overexpression of miR-29b in vivo downregulated STRN4, inhibited cell proliferation, and delayed tumor progression (46). In our study, gga-miR-29b-3p inhibited the invasion and migration of MSB1 cells. Overall, gga-miR-29b-3p exerts a suppressive effect on MD tumor transformation.
Furthermore, we confirmed that gga-miR-29b-3p negatively regulated DNMT3B through directly binding to its 3'-UTR. DNMTs are the predominantly epigenetic modifier genes in mammals and are involved in DNA damage recognition, DNA recombination, and mutation repair (47). In previous studies, miR-29b targeting DNMT3B was also found to exert oncogenic and other effects. Cao et al. [2021] reported that miR-29b promoted cell cycle inhibitor CDKN2B demethylation and transcription by inhibiting DNMT3B activity, induced G1 phase block, inhibited cholangiocarcinoma cell proliferation, and promoted apoptosis (48). Another study reported that miR-29b induced miR-195 promoter demethylation and its re-expression by targeting DNMT3B, thereby inhibiting cell cycle progression and promoting apoptosis in tongue squamous cell carcinoma (TSCC) cell lines (49). In osteoarthritic (OA) cartilage tissue, DNMT3B was shown to induce hypermethylation of specific CpG sites in the miR-29b promoter region which contributed to miR-29b downregulation in OA chondrocytes, both of which were associated with OA-induced chondrocyte apoptosis (50). In our study, transfection of gga-miR-29b-3p mimics resulted in a significant decrease in the expression of DNMT3B at both the transcriptional and translational levels in MSB1. This may be attributed to the complementary pairing of the seed sequence for gga-miR-29b-3p to the 3'-UTR region for DNMT3B. However, further research concerning whether the methylation status of genes is affected by the abnormal regulation of DNMT3B in MSB1 cell is needed to confirm this speculation.
Some researchers have confirmed that DNA methylation is correlated with MD resistance and susceptibility. Yu et al. [2008] reported 2 line-specific DNA transition mutations, CpG→TpG (Chr20:10203733 and 10203778) in MD-susceptible line 72 when compared to the MD-resistance line 63 (51). Tian et al. [2013] found that both global methylation levels and the expression level of DNMT3B were higher in line 72 than in line 63 prior to MDV infection and that the transcriptional expression of DNMT3B was further induced in line 72 after MDV infection (29). These results indicate that the lower methylation in line 63 mostly results from silencing of DNMT3B and DNMT1. We found that interference with DNMT3B inhibited MSB1 cell proliferation and invasion and promoted apoptosis, indicating that gga-miR-29b-3p exert its effects during MD progression by targeting DNMT3B. DNMT3A and DNMT3B are often elevated in various tumors and tumorous cells (52). The role of DNMT3B in many other cancers and its transcriptional regulation by other molecules have been extensively studied (53,54).
MEQ, a 339-amino acid protein, confers an oncogenic property to MDV (55). The colocalization of MEQ with chicken histone deacetylase 1 (HDAC1) and HDAC2 was shown to result in proteasome-dependent degradation (56). The natural killer (NK) cells could be infected with MDV and produced antiviral response, in which MEQ induced NK cell activation (57). Therefore, we investigated whether gga-miR-29b-3p has a suppressive effect on MEQ expression. Both the overexpression of gga-miR-29b-3p and the blockade of DNMT3B decreased the expression of MEQ, suggesting that gga-miR-29b-3p and DNMT3B can regulate the expression of the proto-oncogene MEQ.
Conclusions
Gga-miR-29b-3p presumably acts as a tumor suppressor in MD tumorigenesis by negatively regulating the expression of DNMT3B. Gga-miR-29b-3p exhibited a suppressive effect on cell proliferation, migration, and invasion and facilitated cell apoptosis via the DNMT3B gene. Our study has identified a potential regulatory network of miR-29b-3p and its target gene that regulates the characteristics of MD lymphoma transformation. Our findings further provide information for elucidating the mechanics of MD resistance and susceptibility and for exploring new avenues in the intervention and treatment of MD.
Acknowledgments
Funding: The work was supported in part by the National Natural Science Foundation of China (Nos. 32002160, 32172816, and 31672502), the University Research Project of Anhui Province (No. KJ2020A0081), Anhui Provincial Natural Science Foundation (Nos. 2108085MC117 and 2008085QC140), the Anhui Province Key Research and Development Program Project (No. 202204c06020074), the Anhui Provincial Major Science and Technology Special Program (Nos. 17030701004 and 201903a06020002), the Foundation of Anhui Science and Technology University (No. DKYJ201901), the Graduate Program of the Anhui Provincial Department of Education (No. YJS20210557), and the National Germplasm Center of Domestic Animal Resources.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3519/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3519/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3519/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kaiser P, Underwood G, Davison F. Differential cytokine responses following Marek's disease virus infection of chickens differing in resistance to Marek's disease. J Virol 2003;77:762-8. [Crossref] [PubMed]
- Bertzbach LD, Tregaskes CA, Martin RJ, et al. The Diverse Major Histocompatibility Complex Haplotypes of a Common Commercial Chicken Line and Their Effect on Marek's Disease Virus Pathogenesis and Tumorigenesis. Front Immunol 2022;13:908305. [Crossref] [PubMed]
- Kisielewicz C, Self IA. Canine and feline blood transfusions: controversies and recent advances in administration practices. Vet Anaesth Analg 2014;41:233-42. [Crossref] [PubMed]
- Zai X, Shi B, Shao H, et al. Identification of a Novel Insertion Site HVT-005/006 for the Generation of Recombinant Turkey Herpesvirus Vector. Front Microbiol 2022;13:886873. [Crossref] [PubMed]
- Biggs PM, Nair V. The long view: 40 years of Marek's disease research and Avian Pathology. Avian Pathol 2012;41:3-9. [Crossref] [PubMed]
- Zhang Z, Zhang S, Wang G, et al. Role of microRNA and long non-coding RNA in Marek's disease tumorigenesis in chicken. Res Vet Sci 2021;135:134-42. [Crossref] [PubMed]
- Bertzbach LD, Conradie AM, You Y, et al. Latest Insights into Marek's Disease Virus Pathogenesis and Tumorigenesis. Cancers (Basel) 2020;12:647. [Crossref] [PubMed]
- Reddy SM, Izumiya Y, Lupiani B. Marek's disease vaccines: Current status, and strategies for improvement and development of vector vaccines. Vet Microbiol 2017;206:113-20. [Crossref] [PubMed]
- Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol 2018;141:1202-7. [Crossref] [PubMed]
- Xie M, Ma L, Xu T, et al. Potential Regulatory Roles of MicroRNAs and Long Noncoding RNAs in Anticancer Therapies. Mol Ther Nucleic Acids 2018;13:233-43. [Crossref] [PubMed]
- Chan JJ, Tay Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int J Mol Sci 2018;19:1310. [Crossref] [PubMed]
- Stik G, Dambrine G, Pfeffer S, et al. The oncogenic microRNA OncomiR-21 overexpressed during Marek's disease lymphomagenesis is transactivated by the viral oncoprotein Meq. J Virol 2013;87:80-93. [Crossref] [PubMed]
- Li X, Lian L, Zhang D, et al. gga-miR-26a targets NEK6 and suppresses Marek's disease lymphoma cell proliferation. Poult Sci 2014;93:1097-105. [Crossref] [PubMed]
- Han B, Lian L, Li X, et al. Chicken gga-miR-103-3p Targets CCNE1 and TFDP2 and Inhibits MDCC-MSB1 Cell Migration. G3 (Bethesda) 2016;6:1277-85. [Crossref] [PubMed]
- Zhao C, Li X, Han B, et al. Gga-miR-130b-3p inhibits MSB1 cell proliferation, migration, invasion, and its downregulation in MD tumor is attributed to hypermethylation. Oncotarget 2018;9:24187-98. [Crossref] [PubMed]
- Ding K, Yu ZH, Yu C, et al. Effect of gga-miR-155 on cell proliferation, apoptosis and invasion of Marek's disease virus (MDV) transformed cell line MSB1 by targeting RORA. BMC Vet Res 2020;16:23. [Crossref] [PubMed]
- Zhang Y, Tang N, Luo J, et al. Marek's Disease Virus-Encoded MicroRNA 155 Ortholog Critical for the Induction of Lymphomas Is Not Essential for the Proliferation of Transformed Cell Lines. J Virol 2019;93:e00713-19. [Crossref] [PubMed]
- Lian L, Qu L, Chen Y, et al. A systematic analysis of miRNA transcriptome in Marek's disease virus-induced lymphoma reveals novel and differentially expressed miRNAs. PLoS One 2012;7:e51003. [Crossref] [PubMed]
- Rengaraj D, Lee BR, Lee SI, et al. Expression patterns and miRNA regulation of DNA methyltransferases in chicken primordial germ cells. PLoS One 2011;6:e19524. [Crossref] [PubMed]
- Ponsuksili S, Hadlich F, Reyer H, et al. Genetic background and production periods shape the microRNA profiles of the gut in laying hens. Genomics 2021;113:1790-801. [Crossref] [PubMed]
- Goryo M, Suwa T, Matsumoto S, et al. Serial propagation and purification of chicken anaemia agent in MDCC-MSB1 cell line. Avian Pathol 1987;16:149-63. [Crossref] [PubMed]
- Powell PC, Payne LN, Frazier JA, et al. T lymphoblastoid cell lines from Marek's disease lymphomas. Nature 1974;251:79-80. [Crossref] [PubMed]
- Zhao C, Li X, Han B, et al. Gga-miR-219b targeting BCL11B suppresses proliferation, migration and invasion of Marek's disease tumor cell MSB1. Sci Rep 2017;7:4247. [Crossref] [PubMed]
- Ma Y, Bao J, Zhang Y, et al. Mammalian Near-Infrared Image Vision through Injectable and Self-Powered Retinal Nanoantennae. Cell 2019;177:243-255.e15. [Crossref] [PubMed]
- Li CJ. Flow Cytometry Analysis of Cell Cycle and Specific Cell Synchronization with Butyrate. Methods Mol Biol 2017;1524:149-59. [Crossref] [PubMed]
- Heidari M, Zhang HM, Sharif S. Marek's disease virus induces Th-2 activity during cytolytic infection. Viral Immunol 2008;21:203-14. [Crossref] [PubMed]
- Subramaniam S, Johnston J, Preeyanon L, et al. Integrated analyses of genome-wide DNA occupancy and expression profiling identify key genes and pathways involved in cellular transformation by a Marek's disease virus oncoprotein, Meq. J Virol 2013;87:9016-29. [Crossref] [PubMed]
- Li X, Chiang HI, Zhu J, et al. Characterization of a newly developed chicken 44K Agilent microarray. BMC Genomics 2008;9:60. [Crossref] [PubMed]
- Tian F, Zhan F, VanderKraats ND, et al. DNMT gene expression and methylome in Marek's disease resistant and susceptible chickens prior to and following infection by MDV. Epigenetics 2013;8:431-44. [Crossref] [PubMed]
- Arocho A, Chen B, Ladanyi M, et al. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol Pathol 2006;15:56-61. [Crossref] [PubMed]
- Kurien BT, Scofield RH. Western blotting. Methods 2006;38:283-93. [Crossref] [PubMed]
- Boodhoo N, Gurung A, Sharif S, et al. Marek's disease in chickens: a review with focus on immunology. Vet Res 2016;47:119. [Crossref] [PubMed]
- Hill M, Tran N. miRNA interplay: mechanisms and consequences in cancer. Dis Model Mech 2021;14:dmm047662. [Crossref] [PubMed]
- Botta C, Cucè M, Pitari MR, et al. MiR-29b antagonizes the pro-inflammatory tumor-promoting activity of multiple myeloma-educated dendritic cells. Leukemia 2018;32:1003-15. [Crossref] [PubMed]
- Andrews MC, Cursons J, Hurley DG, et al. Systems analysis identifies miR-29b regulation of invasiveness in melanoma. Mol Cancer 2016;15:72. [Crossref] [PubMed]
- Sur S, Steele R, Shi X, et al. miRNA-29b Inhibits Prostate Tumor Growth and Induces Apoptosis by Increasing Bim Expression. Cells 2019;8:1455. [Crossref] [PubMed]
- Ru P, Steele R, Newhall P, et al. miRNA-29b suppresses prostate cancer metastasis by regulating epithelial-mesenchymal transition signaling. Mol Cancer Ther 2012;11:1166-73. [Crossref] [PubMed]
- Li Y, Cai B, Shen L, et al. MiRNA-29b suppresses tumor growth through simultaneously inhibiting angiogenesis and tumorigenesis by targeting Akt3. Cancer Lett 2017;397:111-9. [Crossref] [PubMed]
- Shin J, Shim HG, Hwang T, et al. Restoration of miR-29b exerts anti-cancer effects on glioblastoma. Cancer Cell Int 2017;17:104. [Crossref] [PubMed]
- Shin J, Shin Y, Oh SM, et al. MiR-29b controls fetal mouse neurogenesis by regulating ICAT-mediated Wnt/β-catenin signaling. Cell Death Dis 2014;5:e1473. [Crossref] [PubMed]
- Nagata S. Apoptosis and Clearance of Apoptotic Cells. Annu Rev Immunol 2018;36:489-517. [Crossref] [PubMed]
- Qu Y, Liao Z, Wang X, et al. EFLDO sensitizes liver cancer cells to TNFSF10-induced apoptosis in a p53-dependent manner. Mol Med Rep 2019;19:3799-806. [Crossref] [PubMed]
- Edlich F. BCL-2 proteins and apoptosis: Recent insights and unknowns. Biochem Biophys Res Commun 2018;500:26-34. [Crossref] [PubMed]
- Cui N, Hu M, Khalil RA. Biochemical and Biological Attributes of Matrix Metalloproteinases. Prog Mol Biol Transl Sci 2017;147:1-73. [Crossref] [PubMed]
- Tabouret E, Boudouresque F, Farina P, et al. MMP2 and MMP9 as candidate biomarkers to monitor bevacizumab therapy in high-grade glioma. Neuro Oncol 2015;17:1174-6. [Crossref] [PubMed]
- Xie Y, Zhao F, Zhang P, et al. miR-29b inhibits non-small cell lung cancer progression by targeting STRN4. Hum Cell 2020;33:220-31. [Crossref] [PubMed]
- Zhang H, Ying H, Wang X. Methyltransferase DNMT3B in leukemia. Leuk Lymphoma 2020;61:263-73. [Crossref] [PubMed]
- Cao K, Li B, Zhang YW, et al. miR-29b restrains cholangiocarcinoma progression by relieving DNMT3B-mediated repression of CDKN2B expression. Aging (Albany NY) 2021;13:6055-65. [Crossref] [PubMed]
- Jia LF, Zheng YF, Lyu MY, et al. miR-29b upregulates miR-195 by targeting DNMT3B in tongue squamous cell carcinoma. Science Bulletin 2016;61:212-9. [Crossref]
- Dou P, He Y, Yu B, et al. Downregulation of microRNA-29b by DNMT3B decelerates chondrocyte apoptosis and the progression of osteoarthritis via PTHLH/CDK4/RUNX2 axis. Aging (Albany NY) 2020;13:7676-90. [Crossref] [PubMed]
- Yu Y, Zhang H, Tian F, et al. An integrated epigenetic and genetic analysis of DNA methyltransferase genes (DNMTs) in tumor resistant and susceptible chicken lines. PLoS One 2008;3:e2672. [Crossref] [PubMed]
- Ehrlich M. DNA hypermethylation in disease: mechanisms and clinical relevance. Epigenetics 2019;14:1141-63. [Crossref] [PubMed]
- Wang L, Wang Z, Huang L, et al. MiR-29b suppresses proliferation and mobility by targeting SOX12 and DNMT3b in pancreatic cancer. Anticancer Drugs 2019;30:281-8. [Crossref] [PubMed]
- Zheng Y, Zhang H, Wang Y, et al. Loss of Dnmt3b accelerates MLL-AF9 leukemia progression. Leukemia 2016;30:2373-84. [Crossref] [PubMed]
- Ennis S, Tai SS, Kihara I, et al. Marek's disease virus oncogene Meq expression in infected cells in vaccinated and unvaccinated hosts. Vet Microbiol 2020;248:108821. [Crossref] [PubMed]
- Liao Y, Lupiani B, Izumiya Y, et al. Marek's disease virus Meq oncoprotein interacts with chicken HDAC 1 and 2 and mediates their degradation via proteasome dependent pathway. Sci Rep 2021;11:637. [Crossref] [PubMed]
- Bertzbach LD, van Haarlem DA, Härtle S, et al. Marek's Disease Virus Infection of Natural Killer Cells. Microorganisms 2019;7:588. [Crossref] [PubMed]
(English Language Editor: J. Gray)


