
Both a hypoxia-inducible EYA3 and a histone acetyltransferase p300 function as coactivators of SIX5 to mediate tumorigenesis and cancer progression
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide and the lifetime risk of developing CRC is nearly 4% in all populations (1,2). The tumorigenesis and progression of CRC arise from different causes such as genetic instability, dysregulated genes (both tumor suppressors and oncogenes), and non-coding RNAs (microRNAs and long non-coding RNAs), the activation and inactivation of signaling pathways involved in tumorigenesis, as well as microenvironment (3,4).
Mammalian genomes encode 6 sine oculis homeobox genes (SIXs) characterized by the presence of the SIX domain (5). The SIX family proteins function as transcription factors to regulate embryogenesis by controlling the renewal and differentiation of progenitor cells (5,6). Following the complement of development, SIXs are typically downregulated (5,6). In recent years, SIX family proteins, especially SIX1, have been identified as being overexpressed in multiple cancers, such as breast cancer, ovarian cancer, prostate cancer, and CRC (7-10). Current data suggests that SIXs can regulate the expression of multiple genes involved in tumorigenesis, cell proliferation, cell migration, invasion, epithelial-to-mesenchymal transition (EMT), and metastasis (7-10). Although SIX proteins contain a DNA-binding domain, they do not harbor an intrinsic transactivation domain (6). Thus, SIXs interact with transcriptional coactivator family members called eyes absent proteins (EYAs, EYA1-4) to activate gene expression (6,7). Like SIXs, EYA members are also dysfunctional after the embryogenesis process but are overexpressed in cancers (11,12). The best demonstrated EYA-SIX partner is EYA1-SIX1, which has been identified to transactivate a series of oncogenes, such as cyclin A1 (CCNA1) (13), cyclin D1 (CCND1) (14), neuropilin 1 (NRP1) (15), and vascular endothelial growth factor C (VEGFC) (16). Except for EYAs, SIX1 can coordinate with a corepressor called dachshund homolog 1 (DACH1) to block p53 gene expression (17).
Gene transcription is controlled by transcriptional complexes assembled by transcription factors and their regulators, such as coactivators [e.g., nuclear receptor coactivators (NOCAs), histone acetyltransferases, and lysine-specific histone demethylase 1A (KDM1A)] and corepressors [e.g., histone deacetylases (HDACs), nuclear receptor corepressors (NCOR), and C-terminal binding proteins (CtBPs)] (18-20). Although the EYA-SIX partners have been well demonstrated to control their target genes in different biological processes, it is still unknown if other transcriptional regulators also participate in the EYA-SIX-controlled transcription.
Currently, the mechanisms that upregulate SIXs and EYAs in tumorigenesis are still being investigated. The hypoxic microenvironment and the activation of hypoxia-inducible factor 1α (HIF-1α) are the two representative features of tumorigenesis and cancer progression (21). Hypoxia has been revealed to stimulate the expression of SIX1 in breast cancer cells (22). However, whether hypoxia induces EYA-SIX partners is still unclear in CRC cells.
Although many studies have discovered that numerous dysregulated signalings and genes may contribute to CRC pathogenesis (3,4), the functions of the EYA-SIX partners and their target genes in CRC tumorigenesis are still obscure. Recently, we explored the expression patterns of six SIX members and four EYA members in cancerous CRC biopsies and CRC cells, and we found that SIX1 and three EYAs (EYA1, EYA3, and EYA4) were upregulated (10). We revealed that the EYA1-SIX1 partner controlled the expression of CCNA1 and transforming growth factor-beta 1 (TGFB1) in the pathogenesis of CRC (10). However, it is still unclear how EYA3 and EYA4 couple with other transcription factors to contribute to CRC tumorigenesis and progression. In the present study, we focused our investigation on revealing the EYA3-associated complex and its downstream target genes in CRC. Our results demonstrated that the hypoxia-induced EYA3 assembled a transcriptional complex with SIX5 and a histone acetyltransferase p300. This complex transactivated the expression of epidermal growth factor receptor (EGFR), vascular endothelial growth factor D (VEGFD), and five matrix metallopeptidases (MMPs) genes by binding to their promoters. Dysfunction of the EYA3-SIX5-p300 complex inhibited the expression of EGFR/VEGFD/MMPs. Our findings reveal the mechanism of how the EYA3-SIX5-p300 complex contributes to CRC pathogenesis and provide a new therapeutic strategy to inhibit CRC cell growth. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2663/rc).
Methods
Cell culture and transfection
Seven CRC cell lines (HT-29, HT55, HCT-15, HCT-116, HCA-24, SW620, and T84) and one noncancerous human colon epithelial cell line (HCEC-1CT) were acquired from the American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Corning, Shanghai, China; #10013CV) containing 10% (v/v) fetal bovine serum (FBS; Corning; #35010CV) at 37 ℃ in a 5% CO2 incubator. Cells under 90% confluence were transfected with plasmids (Table S1) or two independent short hairpin RNAs (shRNAs) targeting each gene (Table S2) following a previous protocol (10).
Tumor xenograft model in mouse
We purchased BALB/c mice from Beijing Vital River Laboratories (Beijing, China) and maintained them in a specific-pathogen-free (SPF) facility with a 12-hour light/dark cycle and free access to water and food. Cells (1×106) were suspended in 100 µL of phosphate-buffered saline (PBS) and then implanted into the right flank of 10-week-old mice (female, weight: 22–24 g, n=10 for each cell line). Tumor volumes were measured every five days. Tumor volumes were determined using the formula: tumor volume = (length × width2)/2. To evaluate the inhibitory effect of the EYA3 inhibitor benzarone (Sigma-Aldrich, Shanghai, China; #B0490000), HT-29 cells (1×106) were injected into the right flank of 10-week-old mice (female, weight: 22–24 g, n=40). After tumor volumes had reached approximately 150 mm3, mice were randomly divided into four subgroups, which were intravenously injected with PBS (control), 50, 100, and 200 mg/kg benzarone (n=10 for each subgroup) at a five-day interval, respectively. At the end of the experiment, mice were sacrificed, and tumors were collected. All mice were grown in the same SPF room. The first author was aware of the group allocation during the experiments. Animal experiments were performed under a project license (No. 2018072HA) granted by the ethics board of West China Hospital of Sichuan University, in compliance with guidelines for the care and use of animals at West China Hospital of Sichuan University.
Data analysis from The Cancer Genome Atlas (TCGA)
Gene expression data and clinical information of 104 CRC patients (50 patients expressed higher levels of EYA3 and 54 patients expressed lower levels of EYA3) were downloaded from TCGA database (https://portal.gdc.cancer.gov/). Approximately 45.2% CRC patients were male and the other 54.8% patients were female. Ages of CRC patients ranged from 47 to 81 years with a median age of 64.3 years. Kaplan-Meier survival curves were used to determine the effects of EYA3 expression levels on overall survival abilities. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell viability and invasion assays
For cell viability, cells were seeded into 96-well plates at a density of nearly 2,500 cells/well. Cell proliferation was determined at different time points (days 0, 1, 2, 3, 4, and 5) using the Cell Counting Kit-8 (CCK-8; Sigma-Aldrich; #96992) according to the guidelines provided by the manufacturer. For cell invasion, nearly 1×105 cells were seeded into the upper chambers of a CultreCoat BME Cell invasion assay kit (R&D systems, Shanghai, China; #3481096K) and incubated at 37 ℃ for 24 hours. The invaded cells in the lower chambers were fixed with methanol and stained with 0.1% (w/v) crystal violet (Sigma-Aldrich; #C6158).
Immunoprecipitation and mass spectrometry
Equal weights of three independent cancerous biopsies were mixed and lysed in a radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich; #R0278) supplemented with a protease inhibitor cocktail (Sigma-Aldrich; #11697498001) for 30 minutes at 4 ℃. Cell lysates were centrifuged at 13,000 rpm for 15 minutes at 4 ℃. The soluble fraction was immunoprecipitated with anti-EYA3 (Proteintech, Wuhan, Hubei, China; #21196-1-AP) associated protein A agarose (Abcam, Shanghai, China; #ab193255). The immunoprecipitates were rinsed six times with RIPA buffer, followed by denaturation in the sodium dodecyl sulfate (SDS) loading buffer. The denatured samples were loaded onto a 10% SDS-polyacrylamide gel electrophoresis (PAGE) gel, followed by silver staining with a kit (Thermo Fisher, Shanghai, China; #24612). The visualized protein bands were destained and digested with sequencing grade trypsin (Thermo Fisher; #90057) overnight at 37 ℃. The eluted peptides were analyzed using a mass spectrometer (Thermo Fisher; #TSQ02-21002) and the obtained data were searched in an in-house Mascot server (Matrix Science, London, UK) against the international protein index with a significance threshold (<0.05).
RNA extraction and quantitative real-time polymerase chain reaction
Total RNA was extracted from cells and biopsies using TRIzol reagent (Thermo Fisher; #15596026). For each sample, 1 µg of total RNA was reverse-transcribed into complementary DNA (cDNA) using the M-MuLV reverse transcriptase (New England Biolabs, Shanghai, China; #M0253S). After dilution for 10-fold, 1 µL cDNA in each sample was used as the template to perform quantitative real-time polymerase chain reaction (qRT-PCR) with the SYBR Green Supermix (Bio-Rad Laboratories, Shanghai, China; #1725270). The primers are all listed in Table S3. The relative messenger RNA (mRNA) level of an individual gene was determined using the 2−ΔΔCT method by normalizing it to Actin.
Western blot assay
Total proteins were extracted using RIPA buffer containing a protease inhibitor cocktail. After centrifuging at 13,000 rpm for 15 minutes at 4 ℃, the soluble cell extracts were used for immunoblots according to a previous protocol (10). The primary antibodies included the following: anti-EYA3 (Proteintech, Wuhan, Hubei, China; #21196-1-AP), anti-SIX5 (Invitrogen, Shanghai, China; #PA5-75417), anti-p300 (Santa Cruz Biotechnology, Shanghai, China; sc-48343), anti-GAPDH (Santa Cruz Biotechnology; #sc-47724), anti-Flag (Sigma-Aldrich; #SAB4200071), and anti-Myc (Invitrogen; #R95125). The horseradish peroxidase (HRP)-conjugated secondary antibodies included the following: anti-mouse immunoglobulin G (IgG) (Sigma-Aldrich; #GENA931) and HRP-conjugated anti-rabbit IgG (Sigma-Aldrich; #GENA934).
Immunohistochemistry (IHC) assay
The IHC assay was performed following a previous method (10). The tissue slices were stained using anti-EYA3. Slides were imaged using an inverted TE 2000 wide-field microscope (Nikon, Tokyo, Japan).
Co-immunoprecipitation (Co-IP) assay
Cells expressing different combinations of plasmids were lysed in RIPA buffer plus 1% protease inhibitor cocktail at 4 ℃ for 2 hours. After centrifuging at 13,000 rpm for 15 minutes at 4 ℃, the supernatant was immunoprecipitated using anti-Flag antibody-conjugated Sepharose beads (Abcam, Shanghai, China; #ab270704) and anti-Myc antibody conjugated Sepharose beads (Sigma-Aldrich; #A7470), respectively. After incubation at 4 ℃ for 2 hours, the beads were rinsed five times with RIPA buffer, followed by boiling with SDS loading buffer and separating in 10% SDS-PAGE gels. The input and output proteins were probed using anti-Myc and anti-Flag.
Chromatin immunoprecipitation (ChIP) assay
Cells were fixed with 1% (w/v) formaldehyde (Sigma-Aldrich; #433284) for 15 minutes, quenched with 125 mM glycine (Sigma-Aldrich; #G8898) for 5 minutes, and washed three times with PBS buffer. Cells were then lysed in the ChIP lysis buffer containing 50 mM HEPES/KOH (pH 7.5), 75 mM NaCl, 1 mM ethylenediamine tetraacetic acid (EDTA), 1% Triton X-100, 0.1% SDS, and 1% protease inhibitor cocktail. Cell lysates were sonicated to reach a 500 bp length of average size, followed by an assay with a ChIP kit (Abcam; #ab500). The input and output DNA were subjected to qRT-PCR analyses to measure the occupancies of EYA3-associated transcriptional complex members on the promoters of genes with the primers listed in Table S4.
Statistical analysis
All experiments were independently repeated in triplicate. Data were presented as the mean ± standard deviation (SD). Statistical analysis was performed using SPSS 19.0 software (IBM Corp., Chicago, IL, USA). The differences between the two groups were analyzed using a Student’s t-test. Figures were generated using GraphPad Prism 8 software (GraphPad Inc., San Diego, CA, USA). Statistical significance was set at a P value less than 0.05.
Results
EYA3 was overexpressed in CRC biopsies and cells
We previously detected the expression patterns of SIXs and EYAs in 24-paired biopsies from CRC patients and discovered that SIX1 and three EYAs (EYA1, EYA3, and EYA4) were upregulated in the cancerous biopsies compared to their adjacent noncancerous tissues (10). We have revealed the functions of EYA1-SIX1 partners in the regulation of genes involved in CRC tumorigenesis. We next aimed to investigate the role of EYA3 and its transcriptional partners. Using the new 20 pairs of biopsies collected from CRC patients, we detected the mRNA levels of EYA3 and we also observed the induction (nearly 2.62±0.33-fold) of EYA3 in the cancerous tissues in comparison to the controls (Figure 1A). Consistently, we also observed the elevation of EYA3 protein levels in the cancerous tissues using IHC and immunoblot assays (Figure 1B-1D).
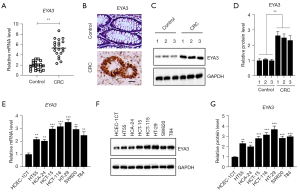
Using one noncancerous cell line HCEC-1CT as a control, we also measured the mRNA and protein levels of EYA3 in seven CRC cell lines (HT-29, HT55, HCT-15, HCT-116, HCA-24, SW620, and T84). Both the real-time quantitative polymerase chain reaction (RT-qPCR) and immunoblot results showed that EYA3 mRNA and protein levels were upregulated in CRC cell lines compared to that in the HCEC-1CT cell line (Figure 1E-1G). Of these seven CRC cells, EYA3 mRNA and protein levels were mostly increased in HT-29 cells (nearly 3.34±0.41-fold, P<0.001) but such increases were the lowest in the HCA-24 cells (nearly 1.91±0.22-fold, P<0.01) (Figure 1E-1G).
EYA3 was induced by hypoxia in vitro
Given that hypoxia is a common feature in tumorigenesis and cancer progression, we next aimed to evaluate if the two hypoxia response transcription factors HIF-1α and HIF-2α were elevated in CRC biopsies. The immunoblot results indicated that both HIF-1α and HIF-2α were increased (HIF-1α 3.14±0.39-fold; HIF-α 2.07±0.26-fold) in three representative pairs of cancerous biopsies compared to their adjacent noncancerous controls (Figure 2A,2B). Using hypoxia-treated HCEC-1CT cells at different time points (0, 3, 6, and 9 h), we found that HIF-1α, HIF-2α, and EYA3 protein levels all gradually increased in a time-dependent manner and they showed similar expression patterns (Figure 2C,2D).
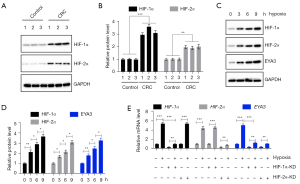
We next evaluated if the expression level of EYA3 was dependent on HIF-1α and HIF-2α. For this purpose, we generated the knockdown (KD) cell lines of both HIF-1α and HIF-2α and then treated cells with or without hypoxia for 9 hours. Hypoxia significantly induced HIF1α, HIF2α, and EYA3 mRNA levels (Figure 2E). Following the depletion of HIF-1α and HIF-2α EYA3 mRNA level was also decreased, and hypoxia treatment only induced the EYA3 mRNA level in HIF-1α-KD and HIF-2α-KD cells to a level comparable to that in non-treated HCEC-1CT cells (Figure 2E). These results revealed that the induction of EYA3 in CRC biopsies and cells was dependent on hypoxia and HIF transcription factors.
The deficiency of EYA3 in CRC cells decreased cell proliferation and invasion
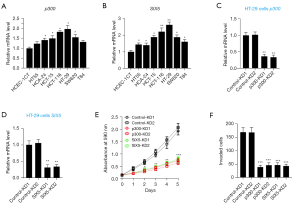
To explore the clinical significance of EYA3, we analyzed the expression level of EYA3 in the clinical dataset of CRC patient samples collected from the TCGA database. The Kaplan-Meier survival curves indicated that the higher expression level of EYA3 was associated with a worse overall survival (OS) rate (Figure 3A).
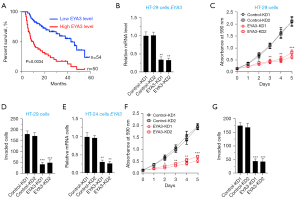
Using HT-29 cells, we generated two independent control-KD cell lines and two EYA3-KD cell lines (Figure 3B). Cell proliferation and invasion assays showed that the deficiency of EYA3 significantly decreased cell viability and invasion rate (Figure 3C,3D, and Figure S1). Similarly, we also created two independent control-KD cell lines and two EYA3-KD cell lines in the HCA-24 background (Figure 3E). We also observed the similar effects of EYA3 depletion on cell proliferation and invasion (Figure 3F,3G, and Figure S1).
EYA3 assembled a transcriptional complex with SIX5 and a histone acetyltransferase p300
To dissect the EYA3-associated transcriptional complex members, we purified the EYA3-associated complex in CRC biopsies using anti-EYA3-conjugated agarose. Mass spectrometry identified 62 EYA3-interacting proteins (Table S5), which included a transcription factor SIX5 and a transcriptional regulator p300 (Table S5). Using the mixture of another three CRC biopsies, we performed an immunoprecipitation assay with anti-EYA3-conjugated agarose, which revealed that SIX5 and p300 could also be pulled down by EYA3 (Figure 4A). To further solidify that EYA3 assembled a complex, we expressed pCDNA3-Flag-EYA3 in HCEC-1CT cells and then conducted an immunoprecipitation assay using anti-Flag-agarose. The results also indicated that EYA3 could pull down both SIX5 and p300 in vitro (Figure 4B).
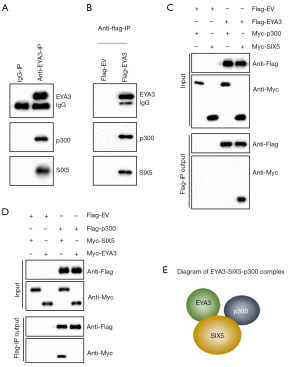
To determine how EYA3 assembled a complex with SIX and p300, we performed Co-IP assays to evaluate the direct interaction between EYA3-p300, EYA3-SIX5, and SIX5-p300. The immunoblot results demonstrated that EYA3 could directly interact with SIX5 but not p300, and SIX5 could directly interact with both EYA3 and p300 (Figure 4C,4D). These results suggested that SIX5 recruited both p300 and EYA3 to form a transcriptional complex (Figure 4E).
Hypoxia promoted the assembly of the EYA3-SIX5-p300 complex
We next evaluated if hypoxia treatment could affect the assembly of the EYA3-SIX5-p300 complex. For this purpose, we treated HCEC-1CT cells with hypoxia for different durations (0, 3, 6, and 9 h), followed by immunoprecipitation using anti-EYA3-conjugated agarose. Consistent with the observation in Figure 2C, the immunoprecipitated EYA3 was gradually increased following the prolongation of hypoxia treatments (Figure S2A). Meanwhile, we found the immunoprecipitated SIX5 and p300 by EYA3 also showed similar patterns to EYA3 (Figure S2A).
Using hypoxia-treated HIF-1α-KD and HIF-2α-KD cells, we performed immunoprecipitation using anti-EYA3-conjugated agarose. The results also indicated that the immunoprecipitated EYA3 in HIF-1α/2α-KD cells and hypoxia-treated HIF-1α/2α-cells were much less than that in the control-KD and hypoxia-treated control-KD cells, respectively (Figure S2B). Similarly, the immunoprecipitated SIX5 and p300 by EYA3 also showed similar patterns to EYA3 in all cells (Figure S2B). These results suggested that hypoxia promoted the assembly of the EYA3-SIX5-p300 complex.
Deficiency of p300 and SIX5 in CRC cells decreased cell proliferation, invasion, and tumor growth
Since EYA3 was elevated in CRC cells, we next measured the expression levels of p300 and SIX5. Using the same seven CRC cell lines and HCEC-1CT control, we found that the p300 mRNA level was slightly increased (from 1.32- to 2.04-fold) in five cell lines (HCA-24, HCT-15, HCT-116, HT-29, and SW620) and the SIX5 mRNA level was similar to the pattern of EYA3 (Figure 5A,5B). We next generated two independent p300-KD and SIX5-KD cell lines in the HT-29 background (Figure 5C,5D). Using these cells, we performed cell proliferation and invasion experiments and the results showed that the depletion of either p300 or SIX5 caused cell growth inhibition and suppressed cell invasion (Figure 5E,5F, and Figure S3). In addition, we also injected control-KD, EYA3-KD, p300-KD, and SIX5-KD cells into mice to generate tumors and then measured tumor volumes at a 5-day interval. The tumor volumes in mice harboring EYA3-KD, p300-KD, and SIX5-KD cells were much smaller than those in mice injected with control-KD cells (Figure S4). Tumor volumes were not obviously different among mice injected with EYA3-KD, p300-KD, and SIX5-KD cells (Figure S4).
The EYA3-SIX5-p300 complex was required for the transregulation of multiple genes involved in tumorigenesis and cancer progression
To identify the downstream target genes of the EYA3-SIX5-p300 complex, we performed a microarray analysis using control-KD1, EYA3-KD1, p300-KD1, and SIX5-KD cells. After analyzing the overlapped genes that were dysregulated in EYA3-KD1, p300-KD1, and SIX5-KD cells compared to the control-KD1 cells, we discovered 29 genes, which included 16 downregulated genes and 13 upregulated genes (Table S6 and Figure 6A). Among them, we observed epidermal growth factor receptor (EGFR), vascular endothelial growth factor D (VEGFD), and five matrix metallopeptidases (MMPs; including MMP3, MMP7, MMP8, MMP21, and MMP26) (Table S6 and Figure 6A). These genes have been previously shown to participate in tumorigenesis and cancer progression.
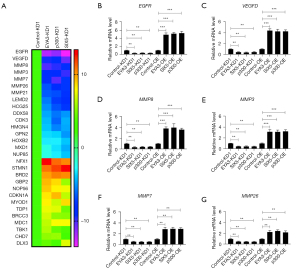
To determine if the microarray data were reliable, we selected 10 representative genes [EGFR, VEGFD, MMP3, MMP7, MMP8, MMP26, RNF5 (ring finger protein 5), TAP2 (transporter 2), STMN1 (stathmin 1), and BRD2 (bromodomain containing 2)] and measured their expression levels in the KD (HT-29 background) and overexpression (OE; HCEC-1CT background) cell lines of EYA3-SIX5-p300 members. Consistent with the microarray results, we observed that the downregulation of EGFR, VEGFD, MMP3, MMP7, MMP8, MMP26, and RNF5 was decreased in the EYA3-KD1, p300-KD1, and SIX5-KD cells compared to the control-KD1 cells (Figure 6B-6G and Figure S5A). In contrast, these seven genes were upregulated in the OE cell lines (Figure 6B-6G and Figure S5A). The other three genes (TAP2, STMN1, and BRD2) were upregulated in the KD cells but downregulated in the OE cells of EYA3-SIX5-p300 members (Figure S5B-S5D).
Given the importance of EGFR, VEGFD, MMP3, MMP7, MMP8, and MMP26 in tumorigenesis and cancer progression, we next measured their expression levels in 20-pairs of CRC biopsies and their adjacent noncancerous tissues. Our results indicated that these genes were all elevated in the cancerous tissues (Figure S6A-S6F).
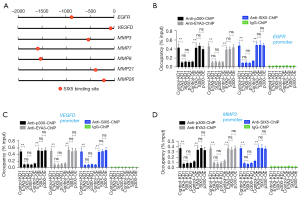
The EYA3-SIX5-p300 complex controlled the expression of EGFR/VEGFD/MMPs through binding to their promoters
To determine if EGFR, VEGFD, MMP3, MMP7, MMP8, MMP21, and MMP26 were the direct targets of the EYA3-SIX5-p300 complex, we analyzed the promoters (2,000 bp length) of these seven genes to identify the SIX5 binding sites using the TCA(A/G)(A/G)TTNC (N represents any nucleotide) consensus sequence. We found that each of these gene promoters contained a SIX5 binding site (Figure 7A). We then performed ChIP assays in the KD and OE cell lines of EYA3-SIX5-p300 members (the same as that used in Figure 6) using anti-EYA3, anti-SIX5, anti-p300, and IgG (negative control). The results indicated that the enrichment of EYA3, SIX5, and p300 on the promoters of EGFR, VEGFD, MMP3, MMP7, MMP8, MMP21, and MMP26 were all decreased in the KD cell lines but increased in the OE cell lines compared to the controls (Figure 7B-7D and Figure S7). Meanwhile, we also detected the expression of MMP13 (its promoter does not contain a SIX5 binding site) in the KD and OE cell lines of EYA3-SIX5-p300 members. We found that the depletion or OE of EYA3-SIX5-p300 members could not change the expression of MMP13 (Figure S8A). The ChIP assay results also supported that the EYA3-SIX5-p300 complex could not bind to the promoter of MMP13 (Figure S8B). These results suggested that EGFR, VEGFD, MMP3, MMP7, MMP8, MMP21, and MMP26 instead of MMP13 were the direct targets of the EYA3-SIX5-p300 complex.
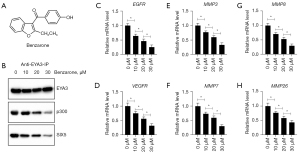
An EYA3 inhibitor benzarone could block the assembly of the EYA3-SIX5-p300 complex and inhibited CRC growth in vitro and in vivo
The results in Figures 3,5 suggested that the EYA3-SIX5-p300 complex might be a potential therapeutic target for CRC treatment. To demonstrate this hypothesis, we treated HT-29 cells with different doses (0, 10, 20, and 30 µM) of an EYA3 inhibitor called benzarone (Figure 8A), followed by an immunoprecipitation assay with anti-EYA3- conjugated agarose. We found that the immunoprecipitated p300 and SIX5 by EYA3 were dose-dependently decreased following benzarone treatments (Figure 8B). Meanwhile, benzarone treatments also caused the dose-dependent suppression of EGFR, VEGFD, MMP3, MMP7, MMP8, and MMP26 (Figure 8C-8H). The ChIP assay results revealed that benzarone treatments caused the dose-dependent decrease in the enrichment of EYA3-SIX5-p300 members on the promoters of EGFR, VEGFD, MMP3, MMP7, MMP8, MMP21, and MMP26 (Figures S9,S10). Consistent with the observation in cells depleted EYA3-SIX5-p300 members, we also found that benzarone treatments resulted in a dose-dependent deduction of cell proliferation (Figure S11A), cell invasion (Figure S11B,S11C), and tumor growth in vivo (Figure S11D).
Discussion
The EYA3 gene functions as a transcriptional partner of SIX transcription factors and the elevation of its expression level has been observed in multiple tumors (11,12). However, three main questions, including the underlying mechanism of EYA3 upregulation, the components of EYA3-associated complex, and the downstream target genes regulated by EYA3-associated complex in the genome-wide, had remained unanswered until now. In the current study, we found that hypoxia induced the expression of EYA3 and promoted the assembly of EYA3-SIX5-p300 in CRC cells. The EYA3-SIX5-p300 complex bound to the promoters of EGFR, VEGFD, MMP3, MMP7, MMP8, MMP21, and MMP26 and induced their expression, thus causing tumorigenesis and promoting cancer progression (Figure 9A). The depletion of EYA3-SIX5-p300 members or the blockage of EYA3-SIX5-p300 assembly by an EYA3 inhibitor benzarone can impair the expression of the downstream target genes, inhibiting CRC cell growth in vitro and in vivo (Figure 9B).
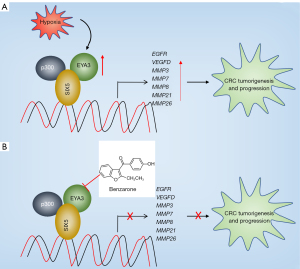
The EYA family proteins function as both transcriptional coactivators and tyrosine phosphatases (11,12). They are involved in multiple biological processes such as cell proliferation, migration, invasion, and metastasis by cooperating with different transcription factors, including SIXs (especially SIX1) and c-Myc (11,12). Currently, studies involved in the interaction between SIXs and EYAs in cancer biology have mainly focused on EYA1-SIX1. We have demonstrated that SIX1 and EYA1 are overexpressed in CRC cells and their interactions activate the expression of CCNA1 and TGFB1 (10). In the current study, we revealed that SIX5 partners with EYA3 and p300 to transactivate a number of genes, including EGFR, VEGFD, MMP3, MMP7, MMP8, MMP21, and MMP26. Of these target genes, EGFR is a strong biomarker of multiple cancer types and its OE can promote solid tumor growth (23). The VEGF-mediated signaling is activated in tumor cells, affecting the function of cancer stem cells, angiogenesis, vascular permeability, and the tumor microenvironment (24,25). Meanwhile, MMPs are involved in matrix degradation and remodeling, tumor neovascularization, and metastasis (26). These seven genes have not been previously identified as targets of either EYAs or SIXs, suggesting that we have identified a new group of direct targets of SIX/EYA partners. Besides these seven genes, we also found that some of the 29 dysregulated genes have been reported to participate in tumorigenesis and cancer progression. For instance, cyclin-dependent kinase 3 (CDK3) can phosphorylate a subunit of activating protein 1 (AP1) and promote the EMT process in CRC cells (27). Cyclin-dependent kinase inhibitor 1A (CDKN1A), also known as p21, is a tumor suppressor (28). The suppression of CDKN1A in tumor cells mainly affects cell cycle progression, leading to uncontrolled cell proliferation (28). However, the promoters of CDK3 and CDKN1A do not contain the SIX5 binding site, suggesting that they may not be the direct target genes of the EYA3-SIX5-p300 complex.
Dynamic changes of histone acetylation and deacetylation affect chromatin structures, increasing or inhibiting gene expression (29). p300 and its homologue CBP serve as scaffolds to recruit transcription factors and other transcriptional regulators, transactivating gene expression (30). One limitation of this study is that we did not investigate the dynamic changes of p300 substrates by proteomics due to the shortage of experimental technology. We will investigate the effects of dynamic changes of acetylation on the expression levels of EGFR/VEGFD/MMPs using p300-specific catalytic inhibitors, bromodomain inhibitors, and p300-knockout cell lines in the future.
Previous publications involved in EYA-SIX-mediated transcription have mainly focused on the interaction between SIXs and EYAs. It is unclear if other transcriptional regulators are involved in the assembly of the EYA-SIX complex. Herein, we discovered that a histone acetyltransferase p300 can be recruited by SIX5, and both EYA3 and p300 are required for the transcription of their target genes. Transcription factors typically recruit p300 to the target gene promoters where p300 acetylates chromatin (31). Although the specific mechanism by which the EYA3-SIX5-p300 complex regulates gene transcription is unclear, we speculate that these three proteins may play their respective roles. One responsibility of SIX5 is binding to its target gene promoters, where it recruits p300 to acetylate chromatin, resulting in the loosened chromatin structure. It further recruits EYA3, which acts as a coactivator to induce the expression of target genes.
Hypoxia microenvironment is common in solid tumors including CRC because the rapid tumor cell proliferation and abnormal tumor blood vessels outstrip the oxygen supply (32). Hypoxia affects numerous tumor biological processes, such as neovascularization, metabolism, metastasis, apoptosis, autophagy, migration, and chemoresistance (33). Each process is controlled by hypoxia-dependent transcriptional programs involving multiple signaling pathways, such as nuclear factor kappa B (NF-κB), phosphoinositide 3-kinases (PI3Ks), and mitogen-activated protein kinase (MAPK) (32,33). An interesting finding in our study is that hypoxia induces EYA3 and promotes the assembly of the EYA3-SIX5-p300 complex. To our knowledge, this is the first report that EYA3 can be induced by hypoxia. The results that depletion of HIF1α and HIF2α decreases the assembly of EYA3-SIX5-p300 complex suggest that HIF inhibitors may be beneficial to CRC therapy by suppressing EGFR, VEGFD, MMP3, MMP7, MMP8, MMP21, and MMP26. However, it is unknown if EYA3 is transcriptionally regulated by the HIF transcription factors. Moreover, EGFR/VEGFD/MMP genes are also involved in the transduction of multiple signalings, such as apoptosis, MAPK/PI3K, and NF-κB (34-36). Thus, more efforts are required to explore the crosstalk between hypoxia signaling and other pathways.
In summary, we demonstrated that p300 and the hypoxia-inducible EYA3 serve as coactivators of SIX5 to transactivate the expression of EGFR, VEGFD, MMP3, MMP7, MMP8, MMP21, and MMP26. Inhibition of EYA3 or specific KD of EYA3-SIX5-p300 members significantly attenuates cell proliferation, invasion, and tumor growth. Our studies uncovered a new mechanism of the assembly of the SIX-associated transcription complex and identified its downstream target genes, providing a new avenue for CRC therapy.
Acknowledgments
Funding: This study was supported by a grant from the National Natural Science Foundation of China (No. 82172909).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-2663/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-2663/dss
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2663/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal experiments were performed under a project license (No. 2018072HA) granted by the ethics board of West China Hospital of Sichuan University, in compliance with guidelines for the care and use of animals at West China Hospital of Sichuan University. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lewandowska A, Rudzki G, Lewandowski T, et al. Title: Risk Factors for the Diagnosis of Colorectal Cancer. Cancer Control 2022;29:10732748211056692. [Crossref] [PubMed]
- Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol 2021;14:101174. [Crossref] [PubMed]
- Al-Joufi FA, Setia A, Salem-Bekhit MM, et al. Molecular Pathogenesis of Colorectal Cancer with an Emphasis on Recent Advances in Biomarkers, as Well as Nanotechnology-Based Diagnostic and Therapeutic Approaches. Nanomaterials (Basel) 2022;12:169. [Crossref] [PubMed]
- Sawicki T, Ruszkowska M, Danielewicz A, et al. A Review of Colorectal Cancer in Terms of Epidemiology, Risk Factors, Development, Symptoms and Diagnosis. Cancers (Basel) 2021;13:2025. [Crossref] [PubMed]
- Jin Y, Zhang M, Li M, et al. SIX1 Activation Is Involved in Cell Proliferation, Migration, and Anti-inflammation of Acute Ischemia/Reperfusion Injury in Mice. Front Mol Biosci 2021;8:725319. [Crossref] [PubMed]
- Meurer L, Ferdman L, Belcher B, et al. The SIX Family of Transcription Factors: Common Themes Integrating Developmental and Cancer Biology. Front Cell Dev Biol 2021;9:707854. [Crossref] [PubMed]
- Blevins MA, Towers CG, Patrick AN, et al. The SIX1-EYA transcriptional complex as a therapeutic target in cancer. Expert Opin Ther Targets 2015;19:213-25. [Crossref] [PubMed]
- Coletta RD, Christensen KL, Micalizzi DS, et al. Six1 overexpression in mammary cells induces genomic instability and is sufficient for malignant transformation. Cancer Res 2008;68:2204-13. [Crossref] [PubMed]
- Behbakht K, Qamar L, Aldridge CS, et al. Six1 overexpression in ovarian carcinoma causes resistance to TRAIL-mediated apoptosis and is associated with poor survival. Cancer Res 2007;67:3036-42. [Crossref] [PubMed]
- Wu J, Huang B, He HB, et al. Two naturally derived small molecules disrupt the sineoculis homeobox homolog 1-eyes absent homolog 1 (SIX1-EYA1) interaction to inhibit colorectal cancer cell growth. Chin Med J (Engl) 2021;134:2340-52. [Crossref] [PubMed]
- Zhang L, Zhou H, Li X, et al. Eya3 partners with PP2A to induce c-Myc stabilization and tumor progression. Nat Commun 2018;9:1047. [Crossref] [PubMed]
- Vartuli RL, Zhou H, Zhang L, et al. Eya3 promotes breast tumor-associated immune suppression via threonine phosphatase-mediated PD-L1 upregulation. J Clin Invest 2018;128:2535-50. [Crossref] [PubMed]
- Coletta RD, Christensen K, Reichenberger KJ, et al. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci U S A 2004;101:6478-83. [Crossref] [PubMed]
- Xu J, Li J, Zhang T, et al. Chromatin remodelers and lineage-specific factors interact to target enhancers to establish proneurosensory fate within otic ectoderm. Proc Natl Acad Sci U S A 2021;118:e2025196118. [Crossref] [PubMed]
- Eisner A, Pazyra-Murphy MF, Durresi E, et al. The Eya1 phosphatase promotes Shh signaling during hindbrain development and oncogenesis. Dev Cell 2015;33:22-35. [Crossref] [PubMed]
- Wang CA, Jedlicka P, Patrick AN, et al. SIX1 induces lymphangiogenesis and metastasis via upregulation of VEGF-C in mouse models of breast cancer. J Clin Invest 2012;122:1895-906. [Crossref] [PubMed]
- Cheng Q, Ning D, Chen J, et al. SIX1 and DACH1 influence the proliferation and apoptosis of hepatocellular carcinoma through regulating p53. Cancer Biol Ther 2018;19:381-90. [Crossref] [PubMed]
- Chen H, Pugh BF. What do Transcription Factors Interact With? J Mol Biol 2021;433:166883. [Crossref] [PubMed]
- Samuels DS, Lybecker MC, Yang XF, et al. Gene Regulation and Transcriptomics. Curr Issues Mol Biol 2021;42:223-66. [PubMed]
- Chen Z. The transrepression and transactivation roles of CtBPs in the pathogenesis of different diseases. J Mol Med (Berl) 2021;99:1335-47. [Crossref] [PubMed]
- Abou Khouzam R, Brodaczewska K, Filipiak A, et al. Tumor Hypoxia Regulates Immune Escape/Invasion: Influence on Angiogenesis and Potential Impact of Hypoxic Biomarkers on Cancer Therapies. Front Immunol 2020;11:613114. [Crossref] [PubMed]
- Zhou H, Blevins MA, Hsu JY, et al. Identification of a Small-Molecule Inhibitor That Disrupts the SIX1/EYA2 Complex, EMT, and Metastasis. Cancer Res 2020;80:2689-702. [Crossref] [PubMed]
- Lawal B, Wang YC, Wu ATH, et al. Pro-Oncogenic c-Met/EGFR, Biomarker Signatures of the Tumor Microenvironment are Clinical and Therapy Response Prognosticators in Colorectal Cancer, and Therapeutic Targets of 3-Phenyl-2H-benzoe1,3-Oxazine-2,4(3H)-Dione Derivatives. Front Pharmacol 2021;12:691234. [Crossref] [PubMed]
- Dakowicz D, Zajkowska M, Mroczko B. Relationship between VEGF Family Members, Their Receptors and Cell Death in the Neoplastic Transformation of Colorectal Cancer. Int J Mol Sci 2022;23:3375. [Crossref] [PubMed]
- Ilson DH. Emerging evidence for VEGF and immune checkpoint inhibition in oesophagogastric cancer. Lancet Gastroenterol Hepatol 2022;7:200-1. [Crossref] [PubMed]
- Cheng T, Chen P, Chen J, et al. Landscape Analysis of Matrix Metalloproteinases Unveils Key Prognostic Markers for Patients With Breast Cancer. Front Genet 2021;12:809600. [Crossref] [PubMed]
- Zheng D, Cho YY, Lau AT, et al. Cyclin-dependent kinase 3-mediated activating transcription factor 1 phosphorylation enhances cell transformation. Cancer Res 2008;68:7650-60. [Crossref] [PubMed]
- Lossaint G, Horvat A, Gire V, et al. Reciprocal regulation of p21 and Chk1 controls the cyclin D1-RB pathway to mediate senescence onset after G2 arrest. J Cell Sci 2022;135:jcs259114. [Crossref] [PubMed]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res 2011;21:381-95. [Crossref] [PubMed]
- Weinert BT, Narita T, Satpathy S, et al. Time-Resolved Analysis Reveals Rapid Dynamics and Broad Scope of the CBP/p300 Acetylome. Cell 2018;174:231-244.e12. [Crossref] [PubMed]
- Maldotti M, Lauria A, Anselmi F, et al. The acetyltransferase p300 is recruited in trans to multiple enhancer sites by lncSmad7. Nucleic Acids Res 2022;50:2587-602. [Crossref] [PubMed]
- Wigerup C, Påhlman S, Bexell D. Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer. Pharmacol Ther 2016;164:152-69. [Crossref] [PubMed]
- Muz B, de la Puente P, Azab F, et al. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl) 2015;3:83-92. [Crossref] [PubMed]
- Mamo M, Ye IC, DiGiacomo JW, et al. Hypoxia Alters the Response to Anti-EGFR Therapy by Regulating EGFR Expression and Downstream Signaling in a DNA Methylation-Specific and HIF-Dependent Manner. Cancer Res 2020;80:4998-5010. [Crossref] [PubMed]
- Goel HL, Mercurio AM. VEGF targets the tumour cell. Nat Rev Cancer 2013;13:871-82. [Crossref] [PubMed]
- Quintero-Fabián S, Arreola R, Becerril-Villanueva E, et al. Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front Oncol 2019;9:1370. [Crossref] [PubMed]


