Association study of the PLXNA4 gene with the risk of Alzheimer’s disease
Introduction
Alzheimer’s disease (AD) has been recognized as the most common form of dementia, affecting more than 13% of senior citizens over 65 years old and as well as 43% population over 85 years old (1,2). Epidemiological studies are still unable to identify one single cause of AD, although age and positive family history are clearly important risk factors (3). The most potent risk factor is apolipoprotein E (APOE) was first elucidated in 1993, and then, then multiple rare mutations in the APP, PSEN1, and PSEN2 genes (participating the encoding process of amyloid precursor protein, presenilin 1, and presenilin 2 respectively) has been discovered through genome-wide association studies (GWAS) (4,5). Swedish twin studies also illustrated that susceptibility alleles may contribute as much as 80% to the risk of the much more common form, late-onset Alzheimer’s disease (LOAD) (6). However, the risk-increasing effects of these genes to AD are much smaller than those of APOE. Thus far, the most well replicated genetic association for LOAD is the variant of the APOE gene (7). APOE Evidence shows that ε4 allele on APOE gene plays major role in the causation of AD (8). Also, it has been estimated that variation at the APOE locus may account for about 50% of LOAD risk (2,7). The ε4 allele frequency in population is of approximately 15%, but is around 40% in LOAD patients (9). Along with APOE ε4, some GWASs and candidate gene investigations have identified at least nine novel risk loci contributing to LOAD, such as CR1, BIN1, CLU, PICALM, CD2AP, CD33, EPHA1, ABCA7 and MS4A cluster (10-15). However, these new discovered gene fail to make full explanation of AD pathogenesis, which indicates that additional risk loci remain to be discovered.
Recent research suggested that Plexin-A 4 (PLXNA4) gene was hitherto demonstrated to participate in the pathogenesis of LOAD for the first time. PLXNA4 is a family member of receptors for trans-membrane, secreted, and glycosylphosphatidylinositol-anchored semaphorins in vertebrates (16) as well as a receptor for secreted Semaphorin3A (SEMA3A) and SEMA6 proteins. This gene plays an important role in semaphorin signaling and axon guidance (17). Accumulation of SEMA3A was previously detected in susceptible areas of the hippocampal neurons during AD progression and co-localized with phosphorylated tau. The staining pattern of SEMA6 presenting in fibers and nerve terminals is also disrupted in brains of patients with AD (18-20). Okada’s team reckoned that PLXNA4 was involved in the precise positioning of oligodendrocyte precursor cells (OPCs) in developing cerebral cortex (21), however, PLXNA4 is not involved in AD through the non-amyloidogenic or amyloidogenic processing of APP (22). These findings elucidate the modulating expression of the individual isoforms, which demonstrates that the genetic variation in PLXNA4 is associated with AD pathogenesis in brain tissue. And evaluate the utility of PLXNA4 as a potential biomarker for AD. Several current studies of PLXNA4 may also provide significant mechanistic evidence linking AD to PD (23,24).
Several single nucleotide polymorphisms (SNPs) including rs277484, rs13057714, rs75460865 and rs13232207 have been reported to increase AD liability by at least 1 rank unit (mean liability on the normalized scale). Research clarifies that rs75460865 and rs13232207 associate with LOAD risk in Caucasians and in Japanese respectively (22). The rs13232207 is located in the PLXNA4 region that encodes the cytoplasmic domain (22); however, there are no similar investigations in Chinese population by now. PLXNA4 gene variant and its frequency in various ethnic groups might be different. We then chose the locus of rs13232207 of PLXNA4 gene to make replication to confirm the potential roles of PLXNA4 in other Asian cohorts. In this study, we firstly implemented the genetic association study on risk locus rs13232207 in PLXNA4 gene in Han Chinese.
Materials and methods
Totally, 978 sporadic LOAD patients (mean age at onset: 75.14±6.07 years; 574 female) and 1,340 healthy control subjects (mean age at examination: 75.50±6.50 years; 740 female) matched for gender and age were included in our research. The LOAD candidates recruited in our study were unrelated Northern Han Chinese citizens in Shandong Province. The AD cases were enrolled from the clinical department of Neurology in Qingdao Municipal Hospital as well as several other hospitals in Shandong Province. The diagnoses of probable AD were defined by at least two neurologists in accordance with the diagnostic criteria of the National Institute of Neurological and Communication Disorders and Stroke-Alzheimer Disease and Related Disorders Association (25). All patients were sporadic cases with those reported a family history of dementia excluded. Their guardians defined the basic characteristics of AD patients, such as onset age and family history. The healthy control subjects were recruited from the Health Examination Center of the Qingdao Municipal Hospital according to the principles described (26). Information authorization was obtained from the all qualified candidates as well as their guardians. The Ethical Committee of Qingdao Municipal Hospital approved our study protocol.
Genomic DNA was extracted from peripheral blood leukocytes of both AD cases and healthy controls, which use the Wizard genomic DNA purification kit (Cat. A1125, Promega, USA). Genotyping of PLXNA4 polymorphism (rs13232207) was accomplished with the SNPscanTM kit providing an accurate and high-throughput method for genotyping 48-192 SNPs simultaneously. SNPscanTM is a patent-pending technology from Shanghai Genesky Biotechnologies Inc., which is developed on double ligation and multiplex fluorescence PCR (27). The primary genotype sequence on the information of the allele-specific ligation-PCR product’s labeling dye color and fragment size is available from the authors upon request.
Hardy-Weinberg equilibrium (HWE) was applied to examine whether the genotype sequence data in both AD cases and healthy controls remain constant from generations in the absence of other evolutionary impacts. After applying the χ2 test, PHWE (P value of HWE test) >0.05 were identified in our research, which means allele frequencies are constant between generations. Comparisons of all variables including differences of alleles and genotype frequencies between AD cases and healthy controls were carried out by χ2 test for categorical variables or Student t-test for continuous variables. And then followed by using multivariate logistic regression adjusting for gender, APOE ε4 status and age to calculate 95% confidence interval (CI) and odds ratios (ORs) under different genetic models (additive, dominant, and recessive). The genetic models were defined as 0 (AA) vs. 1 (Aa + aa) for dominant model, 0 (AA) vs. 1(Aa) vs. 2 (aa) for additive model and 0 (AA + Aa) vs. 1 (aa) for recessive model (a stands for a minor allele and A stands for a major allele). Using logistic regression, the significance of an SNP × APOE interaction was calculated for rs13232207. All statistical analyses were performed through IBM® SPSS® Statistics version 22.0 (SPSS Inc., Chicago, IL, USA). P value <0.05 (based on the SNP analyzed) was considered significant.
Results
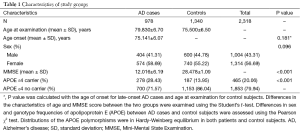
Between LOAD cases and healthy controls, no significant differences were observed regarding sample gender (P=0.096) as well as age (P=0.181). However, the on-set MMSE scores were lower in LOAD patients (12.02±6.19, P<0.001) compared to healthy controls (28.48±1.09, P<0.001). The presence of the APOE ε4 allele raised a higher risk of LOAD in cases than in control subjects (P<0.001; OR =2.449; 95% CI, 1.989–3.014). Detailed information was summarized in Table 1.
Full table
A HWE was applied to examine the genotype sequence data in healthy controls. PHWE =0.067 were identified in HWE test, which means allele frequencies are constant between generations. According to the prevalence of the minor alleles in both groups, the power size of our investigated sample size regarding rs13232207 was over 80% with OR of 1.152 at a significant alpha level of 0.05. Minor allele frequency (MAF) of rs13232207 in cases was 0.238 and in controls was 0.229. The MAFs in both case and control group are in accordance with the MAF in Hapmap, which is 0.333. The OR (95% CI) of locus rs13232207 was 1.048 (0.913–1.202).
In the following statistic process, multivariate logistic regression analysis was used as well as adjusting for sex, age, and APOE ε4 status in total sample. We evaluated the effect of rs13232207 on LOAD risk. The statistic outcome was illustrated clearly in Table 2. All the three models elucidated negative association in the LOAD pathogenesis independently (dominant, OR =1.168, P=0.392; additive, OR =1.087, P=0.233; recessive, OR =1.168, P=0.392). In terms of SNP × APOE interaction, P value shows no significance in the logistic regression with all three models (dominant, P=0.438, additive, P=0.055; recessive, P=0.095). This indicates the T allele has no association with APOE on the risk of LOAD. Careful examination of the genotyping results did not reveal any genotyping errors, which means the minor T allele of rs13232207 might not have any effect on LOAD risk in North Han Chinese with APOE (Table 2).

Full table
Discussion
PLXNA4 represents a functional member of secreting SEMA3A and SEMA6 protein. Previous studies indicated that PLXNA4 was linking to LOAD risk recently. Studies also reported that cysteine repeat modular protein 2 (CRMP2), an intracellular signaling molecule for the semaphorin-plexin signaling pathway, has been observed in neurofibrillary tangles in brains of autopsied AD (28), which supported the association between LOAD and PLXNA4. Rs13232207 encoding cytoplasmic domain is located in the PLXNA4. By genotyping locus rs13232207 in Northern Han Chinese ethnic population, we conducted a specific analysis in a large sample size. According to the outcome of our case-control research, evidence shows the minor T allele play no effect the LOAD risk in the group of APOE ε4 carriers. This outcome is in different with Gyungah Jun’s study in Japanese published in 2014 (22). The MAF of rs13232207 in healthy controls in our research was 0.229. This MAF is higher than in the information (0.0974) obtained from the SNPs database established by the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/SNP). This MAF discrepancy might attribute to the genetic heterogeneity among different ethnic population. In addition, sample sizes in different investigation, population-specific differences as well as gene to gene or gene to environment interactions may also contribute towards this discrepancy in different ethnicity.
PLXNA4 was first proposed to be associated with precise positioning of OPCs in developing cerebral cortex in 2012 (21), Jun’s study then suggested that PLXNA4 do not influence APP processing or Aβ production but its isoform differentially affect Tau protein phosphorylation (22). Through the Tau phosphorylation, disrupted semaphorin-plexin signaling is involved in AD pathogenesis, leading to neurofibrillary tangle formation and neuronal death (29). As these results are based on the transient expression of PLXNA4 molecules, evaluating the influence of PLXNA4 on detailed tau phosphorylation in more physiological conditions is urgent and crucial. Recently a whole exome sequence study identified a rare coding PLXNA4 variant in 2 distantly related individuals with familial Parkinson’s disease (PD) (24). Notably, both PD and AD patients were cognitively deteriorated. Besides, there is a recognized type of PD with dementia. Targeting specific isoforms of PLXNA4 is indeed to clarify the association between PLXNA4 and other cognitive deterioration disease.
There are several caveats in our study. Specimen included in the study was only recruited from Shandong province, which means a limitation of the value in representing Northern Han Chinese. Besides, only 1 locus of PLXNA4 SNPs was analyzed in our research, and thus the lack of an exact replication of other loci may indicate that the initial finding may be a false positive. Third, the MAF is higher than the MAF in the database. However, the discrepancy of MAF can be explained by differences in allele frequencies across populations as well as geographical factors.
To sum up, our investigation failed to support the association of PLXNA4 polymorphism with LOAD risk in a Northern Han Chinese ethnic population. No replicated statistical process was done in the same ethnicity before our efforts. So replication studies are required in the Chinese Han ethnic group. Sufficient large samples, different ethnic populations and the interaction with PLXNA4 and other AD related genes that implicated Tau phosphorylation and SEMA presentation and metabolism are also needed for further investigation to fully assess the relationship between PLXNA4 with LOAD. In the end, genotype-sequencing studies are in an urgent need to identify specific disease related genetic mutations and to examine whether they play a role in other cognitive deterioration related disease as well as AD pathogenesis.
Acknowledgements
Funding: This work was supported by grants from the National Natural Science Foundation of China (No. 81471309, No. 81371406, No. 81571245, and No. 81501103), the Shandong Provincial Outstanding Medical Academic Professional Program, Qingdao Key Health Discipline Development Fund, Qingdao Outstanding Health Professional Development Fund, and Shandong Provincial Collaborative Innovation Center for Neurodegenerative Disorders.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Hebert LE, Scherr PA, Bienias JL, et al. Alzheimer disease in the US population: prevalence estimates using the 2000 census. Arch Neurol 2003;60:1119-22. [Crossref] [PubMed]
- Tan L, Yu JT, Zhang W, et al. Association of GWAS-linked loci with late-onset Alzheimer's disease in a northern Han Chinese population. Alzheimers Dement 2013;9:546-53. [Crossref] [PubMed]
- Breteler MM, Claus JJ, van Duijn CM, et al. Epidemiology of Alzheimer's disease. Epidemiol Rev 1992;14:59-82. [PubMed]
- Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 1993;261:921-3. [Crossref] [PubMed]
- Waring SC, Rosenberg RN. Genome-wide association studies in Alzheimer disease. Arch Neurol 2008;65:329-34. [Crossref] [PubMed]
- Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 2006;63:168-74. [Crossref] [PubMed]
- Ashford JW, Mortimer JA. Non-familial Alzheimer's disease is mainly due to genetic factors. J Alzheimers Dis 2002;4:169-77. [PubMed]
- Wang J, Yu JT, Jiang T, et al. Association of LRRTM3 polymorphisms with late-onset Alzheimer's disease in Han Chinese. Exp Gerontol 2014;52:18-22. [Crossref] [PubMed]
- Bu G. Apolipoprotein E and its receptors in Alzheimer's disease: pathways, pathogenesis and therapy. Nat Rev Neurosci 2009;10:333-44. [Crossref] [PubMed]
- Tan MS, Jiang T, Tan L, et al. Genome-wide association studies in neurology. Ann Transl Med 2014;2:124. [PubMed]
- Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet 2009;41:1088-93. [Crossref] [PubMed]
- Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet 2011;43:429-35. [Crossref] [PubMed]
- Lambert JC, Heath S, Even G, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet 2009;41:1094-9. [Crossref] [PubMed]
- Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet 2011;43:436-41. [Crossref] [PubMed]
- Seshadri S, Fitzpatrick AL, Ikram MA, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 2010;303:1832-40. [Crossref] [PubMed]
- Tamagnone L, Artigiani S, Chen H, et al. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell 1999;99:71-80. [Crossref] [PubMed]
- Suto F, Ito K, Uemura M, et al. Plexin-a4 mediates axon-repulsive activities of both secreted and transmembrane semaphorins and plays roles in nerve fiber guidance. J Neurosci 2005;25:3628-37. [Crossref] [PubMed]
- Good PF, Alapat D, Hsu A, et al. A role for semaphorin 3A signaling in the degeneration of hippocampal neurons during Alzheimer's disease. J Neurochem 2004;91:716-36. [Crossref] [PubMed]
- Hirsch E, Hu LJ, Prigent A, et al. Distribution of semaphorin IV in adult human brain. Brain Res 1999;823:67-79. [Crossref] [PubMed]
- Takegahara N, Kumanogoh A. Immune semaphorins: involvement of semaphorins in various phases of immune responses. Tanpakushitsu Kakusan Koso 2009;54:1101-7. [PubMed]
- Okada A, Tomooka Y. Possible roles of Plexin-A4 in positioning of oligodendrocyte precursor cells in developing cerebral cortex. Neurosci Lett 2012;516:259-64. [Crossref] [PubMed]
- Jun G, Asai H, Zeldich E, et al. PLXNA4 is associated with Alzheimer disease and modulates tau phosphorylation. Ann Neurol 2014;76:379-92. [Crossref] [PubMed]
- Takahashi T, Fournier A, Nakamura F, et al. Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell 1999;99:59-69. [Crossref] [PubMed]
- Schulte EC, Stahl I, Czamara D, et al. Rare variants in PLXNA4 and Parkinson's disease. PLoS One 2013;8:e79145. [Crossref] [PubMed]
- McKhann G, Drachman D, Folstein M, et al. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology 1984;34:939-44. [Crossref] [PubMed]
- Yu JT, Miao D, Cui WZ, et al. Common variants in toll-like receptor 4 confer susceptibility to Alzheimer's disease in a Han Chinese population. Curr Alzheimer Res 2012;9:458-66. [Crossref] [PubMed]
- Xiu L, Lin M, Liu W, et al. Association of DRD3, COMT, and SLC6A4 gene polymorphisms with type 2 diabetes in Southern Chinese: a hospital-based case-control study. Diabetes Technol Ther 2015;17:580-6. [Crossref] [PubMed]
- Cole AR, Knebel A, Morrice NA, et al. GSK-3 phosphorylation of the Alzheimer epitope within collapsin response mediator proteins regulates axon elongation in primary neurons. J Biol Chem 2004;279:50176-80. [Crossref] [PubMed]
- Low LK, Liu XB, Faulkner RL, et al. Plexin signaling selectively regulates the stereotyped pruning of corticospinal axons from visual cortex. Proc Natl Acad Sci U S A 2008;105:8136-41. [Crossref] [PubMed]


