
Long non-coding RNA (FALEC) promotes malignant behaviors of gastric cancer cells by regulating miR-203b/PIM3 axis
Introduction
Gastric cancer (GC) is the most common gastrointestinal malignancy and originates from the lining of the stomach (1). Although the incidence of GC has been largely reduced in the developed countries during the past decades due to the tremendous progress achieved in diagnosis and therapy, GC remains a major threat to human health in the developing world (2). Statistically, more than 200,000 GC-related deaths occurred in China in 2019, which accounted for more than half all GC-related deaths in the world (3). The lack of cancer screening technologies results in a large portion of GC patients being diagnosed at advanced stages, which are characterized by malignant proliferation, distant metastasis, and invasion, all of which are closely correlated with poor prognosis and high mortality (4). Gastrectomy remains the foremost therapeutic option for GC patients, however, it is an impracticable option for GC patients who have extensive invasion or lymphatic metastasis (5). The five-year survival rate of advanced GC patients remains below 5% even after receiving systemic therapy (6). Therefore, it is critical to the mechanism by which GC tumorigenesis operates so that effective GC tumor markers can be developed and the prognosis of GC patients can be improved.
Long non-coding RNAs (lncRNAs) are an important transcript with over 200 nucleotides (7). Numerous studies show the critical role served by lncRNAs as important modulators in multiple cellular biological events involved with cancer, including proliferation, death, migration, and invasion (8,9). In GC, some lncRNAs are upregulated and act as oncogenes, while some lncRNAs exhibit downregulation and act as repressors (10). Previous research shows that focally amplified lncRNA on chromosome 1 (FALEC) affects the occurrence and progression of various human tumors, such as endometrial cancer (11), tongue squamous cell carcinoma (12), colorectal cancer (13), colorectal cancer (13) cervical cancer, melanoma (14), and papillary thyroid carcinoma (15). Recently, FALEC was reported to be upregulated in GC, and its upregulation is closely correlated with lymph node metastasis and TNM stages (16). Besides, previous research also testified that FALEC silencing could prevent GC cell migration and invasion (16). Through the summary of previous literature, we can conclude that FALEC can affect the proliferation, migration, apoptosis and metastasis of cancer cells. Nevertheless, the effect of FALEC on autophagy of cancer cells has not been reported, including GC.
Autophagy is a highly conserved intracellular homeostatic pathway that is responsible for the degradation of damaged intracellular organelles and long-lived proteins (17). Cells can obtain energy during the autophagic process under various cellular stresses, including nutrient-depleted, ischemia, and oxidative stress (18,19). In tumor cells, autophagy may act as a self-defensive mechanism that contributes to tumor cell survival by removing toxins and garbage (20,21). Abnormal autophagic activity also may cause the inappropriate degradation of cell components that are indispensable for tumor c2ell survival, resulting in autophagic cell death (22). Thus, autophagy is crucial in the progression of cancer. Additionally, lncRNAs have a key regulatory effect on tumor cell autophagy, and may be involved in the tumorigenesis of GC (23). In this study, we further investigated the functional roles of FALEC in GC and attempted to determine whether autophagy involves the functional role of FALEC in GC.
Furthermore, growing research confirmed that lncRNAs, as competing endogenous RNA or miRNA sponges, participate in the regulation of cell biological activities (24). Through bioinformatics analysis, we were surprised to find that FALEC and miR-203b have binding sites, indicating that the two may have a relationship of targeted regulation. Recent studies also revealed that miR-203b plays a key role in various cancers (25,26). Besides, through bioinformatics analysis, we also discovered that there were the binding sites between microRNA-203b (miR-203b) and Recombinant Pim-3 Oncogene (PIM3), suggesting that miR-203b may target PIM3. Several studies have also uncovered that PIM3 acts as a carcinogen in various cancer processes (27-29). Therefore, we speculated that miR-203b/PIM3 axis plays a crucial role in the progression of GC. However, the effect of FALEC/miR-203b/PIM3 axis on malignant behavior of GC, especially autophagy, and their targeted regulation relationship are not clear.
In this study, we first verified the effects of FALEC interference on proliferation, migration, invasion, apoptosis and autophagy of GC cells. Subsequently, we further clarified the targeted regulation of FALEC on miR-203b and the targeted regulation of miR-203b on PIM3. In addition, we explored the regulatory roles of FALEC/ Mir-203b /PIM3 axis in the progression of GC. This will provide a theoretical basis for targeted therapy of GC. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1561/rc).
Methods
Tissue collection and cell culture
Twenty pairs of GC and adjacent para-carcinoma tissues were collected from patients who were diagnosed with GC in People’s Hospital of Zhongshan City, then maintained under −80 ℃ until use. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the institutional review board of People’s Hospital of Zhongshan City (No. 2022-003). Informed consent was obtained from each patient. Gastric epithelial cell (GES-1), as well as GC cell lines (AGS, MGC803, and NCI-N87) were all purchased from ATCC and maintained in an incubator with DMEM medium supplemented with 10% fetal bovine serum (FBS) and 1% Penicillin and Streptomycin at 37 ℃.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) assay
Relative expression levels of FALEC, microRNA-203b (miR-203b), and PIM3 mRNA of GC tissues and cells were examined with qRT-PCR. The total RNAs were extracted with TRIzol reagent (Invitrogen, USA) and then RNA concentration was quantified. Next, 3 µg of extracted RNAs were used as a template to generate cDNA with the help of the BestarTM qPCR RT kit (DBI Bioscience, China). The RT-PCR process was conducted on a 7500 Fast Real-Time PCR system (Applied Biosystems, USA) using Power SYBR green master mix (Thermo Fisher Scientific, USA). FALEC and PIM3 expressions were normalized to β-actin, while MiR-203b was normalized to U6. The gene expression was quantified using the 2−ΔΔCT method from 3 independent repetitions (30). Sequences of primers used in the present study are listed in Table 1.
Table 1
| Gene name | Sequence (5'-3') |
|---|---|
| β-actin | Forward: TGGATCAGCAAGCAGGAGTA |
| β-actin | Reverse: TCGGCCACATTGTGAACTTT |
| FALEC | Forward: AGGCAGCAGAACATACAGGA |
| FALEC | Reverse: CCGTTTGAAGTTGCTACCAC |
| MiR-203b | Forward: ACACTCCAGCTGGGTAGTGGTCCTAAACATTT |
| MiR-203b | Reverse: CTCAACTGGTGTCGTGGA |
| MiR-203b | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGTGAAAT |
| PIM3 | Forward: AAGCAGTGACCTCTACCCCTGGTGACC |
| PIM3 | Reverse: CAAATAAATTAAACAATAAATAGCCCC |
| U6 | Forward: CTCGCTTCGGCAGCACA |
| U6 | Reverse: AACGCTTCACGAATTTGCGT |
| U6 | RT: AACGCTTCACGAATTTGCGT |
qRT-PCR, quantitative reverse transcription polymerase chain reaction; FALEC, focally amplified lncRNA on chromosome 1; MiR-203b, microRNA-203b; PIM3, Pim-3 Oncogene.
Transfection of oligonucleotide
Oligonucleotides, including siRNAs against FALEC (si-FALEC), miR-203b mimics, FALEC, and PIM3, as well as their negative controls (NC), were designed and obtained from Integrated Biotech Solutions (Shanghai, China). Oligonucleotide transfection was conducted using Lipofectamine 3000 (Invitrogen) in line with the manufacturer’s protocol.
Western blot
Total proteins were isolated from the processed GC cells using Radio-Immunoprecipitation Assay (RIPA), and protein concentration was determined with the help of a bicinchoninic acid (BCA) kit (Beyotime, China). Protein samples (50 µg) were separated through electrophoresis and then transferred into Poly vinylidene fluoride (PVDF) membranes (Millipore, USA). Afterwards, the membranes were blocked in 5% non-fat milk for 2 h and incubated with the primary antibodies for 8 h against Microtubule-associated protein 1 light chain 3B (LC3B, 1:1000, ab48394, Abcam, UK), Beclin 1 (1:2,000, ab207612, Abcam), P62 (1:500, ab91526, Abcam), Recombinant Pim-3 Oncogene (PIM3, 1:2,000, ab154729, Abcam), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, 1:10,000, ab181602, Abcam). HRP-conjugated donkey-anti-rabbit IgG secondary antibody (Abcam) was used to probe the primary antibody for 2 h. Bands were visualized with the electrochemiluminescence (ECL) system (Thermo Fisher Scientific) and analyzed with Image J software (version 1.50b; National Institutes of Health, Bethesda, MD).
Transmission electron microscopy (TEM)
TEM was used to monitor the autophagy status of treated GC cells. Treated GC cells were collected and washed with PBS and then fixed in 2% glutaraldehyde for 2 h. Cells were then washed with PBS three times and subjected to post-fixation in 1% osmium tetroxide for 1 h and dehydration in graded ethanol for 2 h. TEM observation was conducted with a Leo 912 AB electron microscope.
Immunofluorescence (IF) staining
Treated GC cells were fixed in 4% paraformaldehyde for 30 min and then incubated with TBS containing 5% Tween-20 for 2 h. After the cells were blocked in 10% normal goat serum for 1 h, IF staining was conducted using LC3 primary antibody (1:100, ab62720, Abcam) incubation overnight and subsequent secondary antibody incubation (DAR-546, Abcam) for 2 h. then the cells were dealt with 4’6-diamidino-2-phenylindole (DAPI) for 30 min. Images were photographed using a confocal laser microscope (LSM710, Zeiss, Germany).
Cell proliferation assessment
Cell proliferation was monitored with the cell counting kit-8 (CCK-8) kit (Lianke Bio, China). Cells were harvested after treatment at the exponential proliferation phase and plated onto 96-well plates, followed by 1-, 2-, and 3-days incubation at 37 ℃. CCK-8 solution was added to each well and allowed to incubate for 10 min. The absorbance of each well was measured at 450 nm.
Cell apoptosis assessment
Cell death was estimated with a flow cytometer (BD Bioscience, USA) after staining with Annexin V-FITC Apoptosis Detection Kit (BioVision, USA) and PI following the protocols of manufacturers. Data was analyzed in CellQuest software (version 5.1; Becton-Dickinson and Company).
Cell migration and invasion assessment
Cell migratory and invasive capacities were measured using a transwell assay. Transwell chambers (8 µm, Corning, USA) coated with or without Matrigel were adopted to evaluate the invasive or migratory ability, respectively. Treated GC cells (1×105 cells) suspended in 500 µL DMEM were added into the upper chamber, and 500 µL FBS-included DMEM was added into the lower chamber. After incubating at 37 ℃ for 24 h, non-migrated or non-invaded cells were removed and those cells that migrated or invaded through the membranes were fixed in 4% paraformaldehyde and stained with crystal violet. The number of migratory and invasive cells was counted manually under 10× time magnification.
Prediction of binding sites
RegRNA2.0 (http://regrna2.mbc.nctu.edu.tw/index.html) was applied to predict the binding sites of FALEC to miRNAs (miR-203b); TargetScan analysis (http://targetscan.org) was applied to predict the binding site between miR-1303 and its target genes (PIM3).
Dual-luciferase reporter assay
To verify the interaction between miR-203b and FALEC or PIM3, a dual-luciferase reporter assay was carried out. The wild type and mutant miR-203b binding sequence of FALEC (FALEC-WT, FALEC-MUT) and PIM3 (PIM3-WT, PIM3-MUT) were amplified and inserted into pGL3 luciferase reporter vector to generate recombinant luciferase reporter plasmids. To verify the interaction between miR-203b and FALEC, miR-203b mimics or inhibitors were co-transfected into cells with FALEC-WT or FALEC-MUT, then luciferase intensity assessment was performed with a Dual-Luciferase Reporter Assay System (Promega). Verification of the interaction between miR-203b and PIM3 was carried out in the same way.
Immunohistochemical (IHC) analysis
GC tumor tissues were cut into 5 µm slices after fixation in 4% paraformaldehyde overnight. Slices were incubated with 10% normal donkey serum and then incubated with anti-PIM3 (1:2,000, ab154729, Abcam) primary antibody for 8 h. After washing the slices with PBS, slides were incubated with HRP-conjugated secondary antibody for 1 h. Slices were photographed under a phase-contrast light microscope (Olympus, Japan).
Xenograft tumor proliferation assay
Nude mice (male, 8 weeks old, n=5) obtained from the People’s Hospital of Zhongshan City were kept in a vivarium under a 12 h/12 h day/night circle with free access to food and water. Animal experiments were performed under a project license (No. 2022-003) granted by the ethics committee of People’s Hospital of Zhongshan City, in compliance with People’s Hospital of Zhongshan City guidelines for the care and use of animals. NCI-N87 cells that stably expressed NC or si-FALEC were inoculated subcutaneously into the mice and allowed to proliferate for four weeks. Tumors were dissected after four weeks of inoculation and used for further study. For Hematoxylin and Eosin (H&E) staining, xenograft tumors were fixed in 4% paraformaldehyde and cut into 4-µm slices, followed by H&E staining using the H&E Staining Kit (ab245880, Abcam). And the IHC detection was basically the same as the previous experiment procedure.
Statistical analysis
Data were presented as mean ± standard deviation (SD). The statistical analysis was conducted using SPSS 20.0 software (SPSS, Chicago, IL, USA), and the statistical charts were made using Graphpad (Ver. Prism 8, GraphPad Prism Software, USA). The student’s t-test was adopted to analyze the difference between groups, and One-way analysis of variance was adopted to the difference among over 2 groups. A P value less than 0.05 was considered significant.
Results
FALEC silencing repressed proliferation, migration, and invasion, and induced apoptosis and autophagy in GC cells
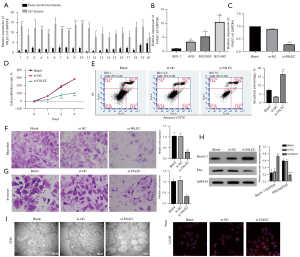
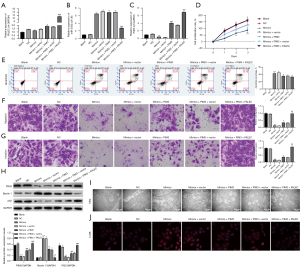
To investigate whether FALEC plays a role during tumorigenesis of GC, we first examined its expression in 20 pairs of GC and matched para-carcinoma tissues samples with qRT-PCR. Results indicated that FALEC was sharply upregulated in 19 GC tissues compared to para-carcinoma ones (Figure 1A). We also found a remarkable upregulation of FALEC in GC cell lines AGS, MGC803, and NCI-N87 compared to that in GES-1 cells (Figure 1B). These findings suggested that FALEC may be involved in the pathogenesis of GC. Furthermore, we estimated the effects of FALEC knockdown on GC cell functions. We first used qRT-PCR analysis to verify the silence effect of FALEC by siRNAs in NCI-N87 cells, and the results showed that FALEC expression was reduced in the si-FALEC group relative to that in the si-NC group (Figure 1C). Then we found that FALEC knockdown dramatically reduced NCI-N87 cell proliferation (Figure 1D), and that FALEC knockdown increased the apoptosis rate of NCI-N87 cells (Figure 1E). Additionally, we found that FALEC knockdown caused a prominent repressive effect on the migratory and invasive capacities of NCI-N87 cells (Figure 1F,1G). Also, compared to the si-NC group, Beclin 1 expression was increased, while P62 expression was significantly decreased in the FALEC-silenced group (Figure 1H). Moreover, results from TEM indicated a substantial increase in the accumulation of autophagic vesicles in si-FALEC transfected NCI-N87 cells (Figure 1I). A prominent upregulation of LC3B was also identified in the si-FALEC group relative to the si-NC group (Figure 1J). Our data indicated that FALEC knockdown promoted GC cell apoptosis and autophagy, while it repressed GC cell proliferation, migration, and invasion in vitro.
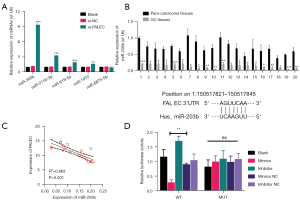
MiR-203b was sponged by FALEC
Then we screened the potential target miRNAs of FALEC using the starBase database. Through qRT-PCR analysis, we found that miRNAs, including miR-203b, miR-2116-3p, miR-619-5p, miR-1972, and miR-6875-5p, were prominently downregulated in FALEC-silenced NCI-N87 cells, especially miR-203b (Figure 2A). miR-203b was selected for further study. Next, qRT-PCR analysis of miR-203b showed that it was downregulated in 20 GC tissues (Figure 2B). Moreover, we found a negative correlation between the expression of FALEC and miR-203b in GC (Figure 2C). Next, we verified the physical interaction between FALEC and miR-203b. The luciferase activity of GC cells driven by FALEC-WT was sharply attenuated by the transfection of miR-203b mimics (Figure 2D). Our findings show that miR-203b can target FALEC in GC.
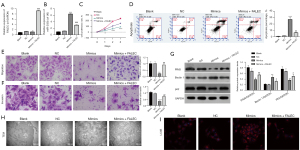
FALEC relieved the regulatory effects of miR-203b overexpression on GC cell functions
To determine whether miR-203b participates in the regulation of FALEC on GC progression, NCI-N87 cells were first transfected with miR-203b mimics and FALEC plasmid. As presented in Figure 3A,3B, transfection of miR-203b mimics upregulated miR-203b, then overexpression of FALEC upregulated FALEC and downregulated miR-203b in miR-203b-overexpressed NCI-N87 cells. Subsequently, CCK-8 data showed that, compared to the NC group, cell proliferation in the miR-203b group was significantly repressed, while co-transfection of miR-203b and FALEC partially reversed the inhibitory effects of miR-203b mimics on cell proliferation (Figure 3C). Results of apoptosis analysis showed that transfection of miR-203b mimics caused an increase of cell apoptosis, while this phenomenon was partially abolished by FALEC (Figure 3D). Moreover, the transwell assay indicated a significant inhibitory effect on the migratory and invasive capacities in the miR-203b group, which also could be abrogated by FALEC (Figure 3E,3F). Meanwhile, we estimated the change of cell autophagy. PIM3 and P62 expressions decreased, while Beclin 1 was upregulated in miR-203b-overexpressed NCI-N87 cells, and these dysregulations could be reversed by FALEC overexpression (Figure 3G). A significant increase of autophagic vesicles was also observed in the miR-203b-overexpressed NCI-N87 cells, while this increase could be abolished by FALEC overexpression (Figure 3H). The data indicated that miR-203b overexpression caused a prominent induction on autophagy (LC3B), while FALEC overexpression could abrogate this effect (Figure 3I). Therefore, FALEC reversed the inhibitory effects of miR-203b overexpression on GC cell functions in vitro.
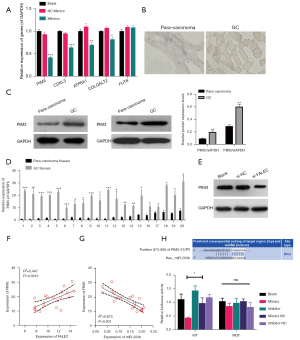
PIM3 was targeted by miR-203b
With TargetScan, we found five genes, including PIM3, CDKL3, ATP8A1, COLGALT2, and FUT8, that might be the potential target genes of miR-203b. To screen for the most effective target genes, qRT-PCR was conducted in miR-203b-overexpressed NCI-N87 cells. The results showed that overexpression of miR-203b could cause prominent downregulation of PIM3, CDKL3, ATP8A1, and COLGALT2 in NCI-N87 cells, especially PIM3 (Figure 4A). PIM3 was selected and verified in subsequent experiments. First, IHC data showed that PIM3 was upregulated in GC tissues (Figure 4B). Second, PIM3 also indicated a prominent upregulation of PIM3 in GC tissues compared to para-carcinoma tissues (Figure 4C,4D). Meanwhile, we found that the silence of FALEC downregulated PIM3 in NCI-N87 cells (Figure 4E). Furthermore, a positive correlation was observed between PIM3 and FALEC expression (Figure 4F), and a negative correlation was identified between PIM3 and miR-203b expression (Figure 4G). Physical interaction between miR-203b and PIM3 was also verified. Luciferase activity of GC cells driven by PIM3-WT was attenuated by miR-203b mimics and was enhanced by miR-203b inhibitor (Figure 4H). Thus, these results suggested that PIM3 was targeted by and inversely correlated with miR-203b in GC.
FALEC overexpression induced proliferation, migration, and invasion, and prevented apoptosis and autophagy through the miR-203b/PIM3 axis
We further determined whether PIM3 participates in the regulatory effects of FALEC/miR203b on the malignant behaviors of GC. qRT-PCR results uncovered that the transfection of miR-203b mimics could downregulate FALEC and PIM3, while overexpression of PIM3 and FALEC could reverse the levels of FALEC and PIM3 mediated by miR-203b overexpression in NCI-N87 cells; we also discovered that overexpressed-miR-203b, which was induced by miR-203b mimics, could be dramatically weakened by overexpression of FALEC in NCI-N87 cells (Figure 5A-5C). Meanwhile, And the results of functional experiments displayed that the repressive effects of miR-203b overexpression on cell proliferation, migration, and invasion were reversed by PIM3 overexpression. Similarly, our data also verified that the promotive effects of miR-203b overexpression on cell apoptosis also could be reversed by PIM3 overexpression. Moreover, the rescue effects of PIM3 also could be further enhanced by FALEC overexpression (Figure 5D-5G). Next, the PIM3 overexpression could reverse the downregulation of PIM3 and P62 and the upregulation of Beclin1 induced by miR-203b overexpression, while this phenomenon could be blocked by FALEC overexpression (Figure 5H). Then TEM results indicated that the stimulative effect of miR-203b overexpression on autophagosomes could be relieved by PIM3 overexpression, and this phenomenon also could be abolished by FALEC overexpression (Figure 5I). This conclusion was further supported by the IF staining of LC3B (Figure 5J). Consequently, we revealed that FALEC could prominently accelerate GC progression through the miR-203b/PIM3 axis.
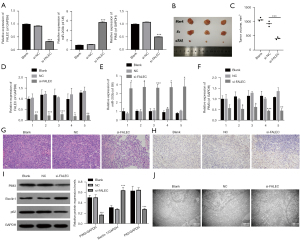
FALEC knockdown inhibited growth and induced autophagy in vivo
To further determine the role of FALEC in GC tumor proliferation and autophagy in vivo, a xenograft tumor assay was performed. NCI-N87 cells were transfected with si-FALAEC and the control (NC), and the expression changes of FALEC, miR-203b, and PIM3 were identified through qRT-PCR assay. As displayed in Figure 6A, silence of FALAEC reduced FALEC and PIM3 expressions and elevated miR-203b expression in NCI-N87 cells (P<0.001). Next, FALEC-silenced NCI-N87 cells were subcutaneously inoculated into the left flank of nude mice and allowed to proliferation for four weeks (Figure 6B). We found that the volume of tumors was smaller in the si-FALEC group than in the NC group (Figure 6C). Moreover, we detected the expression of FALEC, miR-203b, and PIM3 in the xenograft tumors formed by si-FALEC transfected cells with qRT-PCR. Results indicated that FALEC and PIM3 were downregulated, while miR-203b was upregulated in the si-FALEC transfected group compared to the NC group (Figure 6D-6F). Next, H&E staining results indicated that the tumors formed by si-FALEC transfected cells showed well differentiation (Figure 6G). Immunohistochemical analysis showed downregulation of PIM3 in xenograft tumors formed by si-FALEC transfected cells (Figure 6H). Results from western blot analysis indicated that PIM3 and P62 expression was decreased, while Beclin 1 expression was increased in the si-FALEC group (Figure 6I), indicating that autophagy in the si-FALEC group was promoted. Meanwhile, the promotion of autophagy was further supported by the TEM results (Figure 6J). These findings indicated that FALEC knockdown regulated autophagy, miR-203b, and PIM3 in vivo.
Discussion
FALEC, a lncRNA transcript with 566 nucleotides located at 1q21.2, is an important oncogene in multiple human cancers, including prostate cancer, papillary thyroid carcinoma, and melanoma (14,15,31). However, it was found to be downregulated in tongue squamous cell carcinoma (TSCC), and exhibited a repressive effect on TSCC cell proliferation and metastasis by silencing extracellular matrix protein 1 (ECM1) through EZH2 (12). Therefore, its expression level and biological function in different types of cancer may be different. Recent research showed that FALEC was increased in GC, and that knockdown of FALEC inhibited GC progression by impairing ECM1 expression (16). Consistently, in our study, we confirmed the upregulation of FALEC in GC tissues and cell lines, and knockdown of FALEC exhibited a repressive effect on GC progression in vitro and in vivo. Moreover, we found that FALEC knockdown could induce GC cell autophagy. Regarding the mechanism, FALEC could regulate the expression of PIM3 by sponging miR-203b in GC and by regulating the autophagy, proliferation, apoptosis, migration, and invasion of GC cells.
LncRNAs share a general functionality in their capacity to regulate gene expression by sponging miRNAs in a phenomenon defined as competing for endogenous RNA (ceRNA) theory (32,33). The hypothesis of ceRNA is a common mechanism of lncRNA in regulating tumor progression (34). MiRNAs have functions similar to oncogenes or tumor suppressor genes and are closely related to the development process of human tumors (35,36). Recent studies have identified that miRNAs are relevant to the occurrence, development, and metastasis of GC (37,38). Therefore, we screened the target miRNAs of FALEC with the starBase database. The new studies confirmed that miR-203b could act as a tumor inhibitor in a variety of cancers, including Gastric Cardia Adenocarcinoma (39), esophageal squamous cell carcinoma (25), skin cancer (40), and colorectal cancer (41). After screening, miR-203b was identified as the most promising. We further found that miR-203b decreased in GC. Meanwhile, miR-203b overexpression could prevent proliferation, migration, and invasion, and induce apoptosis and autophagy, which could also be abolished by the overexpression of FALEC in GC cells. Furthermore, we also verified the physical interaction between miR-203b and FALEC. Therefore, we found that FALEC accelerated the malignant biological properties of GC cells by targeting miR-203b.
Research also shows that miRNAs can directly degrade mRNA or prevent translation by negatively regulating target genes and affecting a number of cell activities, such as cell growth, proliferation, apoptosis, autophagy, inflammation, invasion, and metastasis (36,42,43). Additionally, we also predicted the potential target genes of miR-203b using TargetScan, and PIM3 was identified to be the potential target gene of miR-203b. PIM kinase family is a group of calcium/calmodulin regulated kinase (CAMP) families whose members (PIM-1, PIM-2, PIM-3) are highly homologous in the kinase domain (44). Research shows that PIM family kinases can significantly induce cell survival and proliferation (45,46). Recent studies indicate that PIM-3 is abnormally expressed in a variety of cancers, including prostate cancer (47), colorectal cancer (48), pancreatic cancer (49), rectal cancer (50), and melanoma (51). Therefore, PIM-3 might be a novel target for anti-tumor therapy. Nevertheless, the role of miR-203b and PIM3 in GC tumorigenesis has not been reported yet. In our study, we found that PIM3 was a targeted gene of miR-203b and could be negatively regulated by miR-203b in GC. We also found that the miR-203b/PIM3 axis was involved in the regulation of FALEC on the autophagy, proliferation, migration, invasion, and apoptosis in GC. In summary, FALEC/miR-203b/PIM3 works as an axis to regulate the malignant activities of GC cells.
Tumor distant metastasis usually indicates advanced progression and poor overall survival rate in patients with GC (52). The tumor metastasis process is extremely complex, involving multiple pathological activities, including angiogenesis, autophagy, and epithelial-to-mesenchymal transition (53). Autophagy is an evolutionarily highly conserved metabolic pathway involving multiple steps, which can maintain the homeostasis of the intracellular environment by degrading damaged proteins, cellular metabolites, and diseased organelles. LC3 and P62, as the key markers of autophagy, are frequently applied to evaluate autophagy levels. Beclin-1, as a tumor suppressor gene, is affected by autophagy during cancer progression. Moreover, induction of autophagy is associated with the prevention of GC progression in multiple studies (54,55), which is consistent with our conclusions.
In conclusion, the results from the in vitro and in vivo experiments suggested that FALEC accelerates GC cell malignant behaviors by regulating the miR-203b/PIM3 axis. These findings might provide a potential therapeutic axis for GC treatment.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1561/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1561/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1561/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the institutional review board of People’s Hospital of Zhongshan City (No. 2022-003). Informed consent was obtained from each patient. Animal experiments were performed under a project license (No. 2022-003) granted by the ethics committee of People’s Hospital of Zhongshan City, in compliance with People’s Hospital of Zhongshan City guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Strong VE. Progress in gastric cancer. Updates Surg 2018;70:157-9. [Crossref] [PubMed]
- Venerito M, Vasapolli R, Rokkas T, Malfertheiner P. Gastric cancer: epidemiology, prevention, and therapy. Helicobacter 2018;23:e12518. [Crossref] [PubMed]
- Strong VE, Wu AW, Selby LV, et al. Differences in gastric cancer survival between the U.S. and China. J Surg Oncol 2015;112:31-7. [Crossref] [PubMed]
- Digklia A, Wagner AD. Advanced gastric cancer: Current treatment landscape and future perspectives. World J Gastroenterol 2016;22:2403-14. [Crossref] [PubMed]
- Song Z, Wu Y, Yang J, et al. Progress in the treatment of advanced gastric cancer. Tumour Biol 2017;39:1010428317714626. [Crossref] [PubMed]
- Ohtsu A, Ajani JA, Bai YX, et al. Everolimus for previously treated advanced gastric cancer: results of the randomized, double-blind, phase III GRANITE-1 study. J Clin Oncol 2013;31:3935-43. [Crossref] [PubMed]
- Weidle UH, Birzele F, Kollmorgen G, et al. Long Non-coding RNAs and their Role in Metastasis. Cancer Genomics Proteomics 2017;14:143-60. [Crossref] [PubMed]
- Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci 2017;108:1927-33. [Crossref] [PubMed]
- Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017;36:5661-7. [Crossref] [PubMed]
- Lin MT, Song HJ, Ding XY. Long non-coding RNAs involved in metastasis of gastric cancer. World J Gastroenterol 2018;24:3724-37. [Crossref] [PubMed]
- Zheng QH, Shi L, Li HL. FALEC exerts oncogenic properties to regulate cell proliferation and cell-cycle in endometrial cancer. Biomed Pharmacother 2019;118:109212. [Crossref] [PubMed]
- Jia B, Xie T, Qiu X, et al. Long noncoding RNA FALEC inhibits proliferation and metastasis of tongue squamous cell carcinoma by epigenetically silencing ECM1 through EZH2. Aging (Albany NY) 2019;11:4990-5007. [Crossref] [PubMed]
- Jiang H, Liu H, Jiang B. Long non-coding RNA FALEC promotes colorectal cancer progression via regulating miR-2116-3p-targeted PIWIL1. Cancer Biol Ther 2020;21:1025-32. [Crossref] [PubMed]
- Ni N, Song H, Wang X, et al. Up-regulation of long noncoding RNA FALEC predicts poor prognosis and promotes melanoma cell proliferation through epigenetically silencing p21. Biomed Pharmacother 2017;96:1371-9. [Crossref] [PubMed]
- Xiao X, Li L, Cui JC, et al. LncRNA FALEC promotes proliferation, migration, and invasion of PTC cells through regulating Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci 2020;24:4361-7. [PubMed]
- Wu H, Qiao F, Zhao Y, et al. Downregulation of Long Non-coding RNA FALEC Inhibits Gastric Cancer Cell Migration and Invasion Through Impairing ECM1 Expression by Exerting Its Enhancer-Like Function. Front Genet 2019;10:255. [Crossref] [PubMed]
- Cao Y, Luo Y, Zou J, et al. Autophagy and its role in gastric cancer. Clin Chim Acta 2019;489:10-20. [Crossref] [PubMed]
- White E, Mehnert JM, Chan CS. Autophagy, Metabolism, and Cancer. Clin Cancer Res 2015;21:5037-46. [Crossref] [PubMed]
- Wang P, Shao BZ, Deng Z, et al. Autophagy in ischemic stroke. Prog Neurobiol 2018;163-164:98-117. [Crossref] [PubMed]
- Yang X, Yu DD, Yan F, et al. The role of autophagy induced by tumor microenvironment in different cells and stages of cancer. Cell Biosci 2015;5:14. [Crossref] [PubMed]
- Maes H, Rubio N, Garg AD, et al. Autophagy: shaping the tumor microenvironment and therapeutic response. Trends Mol Med 2013;19:428-46. [Crossref] [PubMed]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008;132:27-42. [Crossref] [PubMed]
- Xin L, Zhou Q, Yuan YW, et al. METase/lncRNA HULC/FoxM1 reduced cisplatin resistance in gastric cancer by suppressing autophagy. J Cancer Res Clin Oncol 2019;145:2507-17. [Crossref] [PubMed]
- Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol 2016;1402:271-86. [Crossref] [PubMed]
- Liu Y, Dong Z, Liang J, et al. Methylation-mediated repression of potential tumor suppressor miR-203a and miR-203b contributes to esophageal squamous cell carcinoma development. Tumour Biol 2016;37:5621-32. [Crossref] [PubMed]
- Yalçınkaya B, Güzel E, Taştekin D, et al. Role of mir-33a, mir-203b, mir361-3p, and mir-424 in hepatocellular carcinoma Turk J Med Sci 2021;51:638-43. [Crossref] [PubMed]
- Luo H, Sun R, Zheng Y, et al. PIM3 Promotes the Proliferation and Migration of Acute Myeloid Leukemia Cells. Onco Targets Ther 2020;13:6897-905. [Crossref] [PubMed]
- Qi Q, Pan Y, Han S, et al. PIM3 Functions as Oncogenic Factor and Promotes the Tumor Growth and Metastasis in Colorectal Cancer. Anat Rec (Hoboken) 2019;302:1552-60. Retraction in: Anat Rec (Hoboken) 2021;304:2897. [Crossref] [PubMed]
- Zhou Y, Zhou YN, Liu SX, et al. Effects of PIM3 in prognosis of colon cancer. Clin Transl Oncol 2021;23:2163-70. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Zhao R, Sun F, Bei X, et al. Upregulation of the long non-coding RNA FALEC promotes proliferation and migration of prostate cancer cell lines and predicts prognosis of PCa patients. Prostate 2017;77:1107-17. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. [Crossref] [PubMed]
- Guo LL, Song CH, Wang P, et al. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol 2015;21:11680-7. [Crossref] [PubMed]
- Cortez MA, Anfossi S, Ramapriyan R, et al. Role of miRNAs in immune responses and immunotherapy in cancer. Genes Chromosomes Cancer 2019;58:244-53. [Crossref] [PubMed]
- Xiong Q, Su H. MiR-325-3p functions as a suppressor miRNA and inhibits the proliferation and metastasis of glioma through targeting FOXM1. J Integr Neurosci 2021;20:1019-28. [Crossref] [PubMed]
- Riquelme I, Letelier P, Riffo-Campos AL, et al. Emerging Role of miRNAs in the Drug Resistance of Gastric Cancer. Int J Mol Sci 2016;17:424. [Crossref] [PubMed]
- Yang W, Ma J, Zhou W, et al. Molecular mechanisms and theranostic potential of miRNAs in drug resistance of gastric cancer. Expert Opin Ther Targets 2017;21:1063-75. [Crossref] [PubMed]
- Liu W, Dong Z, Liang J, et al. Downregulation of Potential Tumor Suppressor miR-203a by Promoter Methylation Contributes to the Invasiveness of Gastric Cardia Adenocarcinoma. Cancer Invest 2016;34:506-16. [Crossref] [PubMed]
- Bhalla S, Kaur H, Dhall A, et al. Prediction and Analysis of Skin Cancer Progression using Genomics Profiles of Patients. Sci Rep 2019;9:15790. [Crossref] [PubMed]
- Yang Z, An Y, Wang N, et al. LINC02595 promotes tumor progression in colorectal cancer by inhibiting miR-203b-3p activity and facilitating BCL2L1 expression. J Cell Physiol 2020;235:7449-64. [Crossref] [PubMed]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. [Crossref] [PubMed]
- Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol 2018;141:1202-7. [Crossref] [PubMed]
- Asati V, Mahapatra DK, Bharti SK. PIM kinase inhibitors: Structural and pharmacological perspectives. Eur J Med Chem 2019;172:95-108. [Crossref] [PubMed]
- Fan X, Xie Y, Zhang L, et al. Effect of Pim-3 Downregulation on Proliferation and Apoptosis in Lung Adenocarcinoma A549 Cells. Ann Clin Lab Sci 2019;49:770-6. [PubMed]
- Wu F, Huang W, Yang L, et al. MicroRNA-101-3p regulates gastric cancer cell proliferation, invasion and apoptosis by targeting PIM 1 expression. Cell Mol Biol (Noisy-le-grand) 2019;65:118-22. [Crossref] [PubMed]
- Qu Y, Zhang C, Du E, et al. Pim-3 is a Critical Risk Factor in Development and Prognosis of Prostate Cancer. Med Sci Monit 2016;22:4254-60. [Crossref] [PubMed]
- Zhou Z, Zhang R, Wang R, et al. Expression of Pim-3 in colorectal cancer and its relationship with prognosis. Tumour Biol 2016;37:9151-6. [Crossref] [PubMed]
- Li T, Wang Z, Hou YF, et al. Pim-3 Regulates Stemness of Pancreatic Cancer Cells via Activating STAT3 Signaling Pathway. J Cancer 2017;8:1530-41. [Crossref] [PubMed]
- Zhang RX, Zhou ZG, Lu SX, et al. Pim-3 as a potential predictor of chemoradiotherapy resistance in locally advanced rectal cancer patients. Sci Rep 2017;7:16043. [Crossref] [PubMed]
- Liu J, Qu X, Shao L, et al. Pim-3 enhances melanoma cell migration and invasion by promoting STAT3 phosphorylation. Cancer Biol Ther 2018;19:160-8. [Crossref] [PubMed]
- Jin X, Zhu Z, Shi Y. Metastasis mechanism and gene/protein expression in gastric cancer with distant organs metastasis. Bull Cancer 2014;101:E1-12. [Crossref] [PubMed]
- Su Z, Yang Z, Xu Y, et al. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol Cancer 2015;14:48. [Crossref] [PubMed]
- Li GM, Li L, Li MQ, et al. DAPK3 inhibits gastric cancer progression via activation of ULK1-dependent autophagy. Cell Death Differ 2021;28:952-67. [Crossref] [PubMed]
- Lv GB, Wang TT, Zhu HL, et al. Vortioxetine induces apoptosis and autophagy of gastric cancer AGS cells via the PI3K/AKT pathway. FEBS Open Bio 2020;10:2157-65. [Crossref] [PubMed]
(English Language Editor: C. Mullens)


