
The utility of sputum supernatant as an alternative liquid biopsy specimen for next-generation sequencing-based somatic variation profiling
Introduction
Genomic profiling is a valuable diagnostic assay in enabling opportunities for individualized treatment of cancer (1). DNA-based next-generation sequencing (NGS) using hybrid capture of tens to hundreds of cancer-related genes enables the simultaneous detection of clinically relevant sequence variations for various purposes, including diagnosis, prognosis, and treatment selection. NGS is now recommended by the European Society for Medical Oncology (ESMO) for routine use on tumor or plasma samples from patients with metastatic cancers, including non-small cell lung cancer (NSCLC) (2). Although tissue biopsy is the preferred specimen for genomic profiling, circulating cell-free DNA (cfDNA) isolated from liquid biopsy specimens are considered good alternative sources of tumor DNA, particularly when tissue samples are limited or inaccessible (3,4). Compared with tissue biopsy, liquid biopsy specimens are more accessible and less invasive, making them a better sample type for clinical applications that require repeated sampling, such as for treatment monitoring (3,4). The rapid technological advancements in molecular assays have led to improvement of the diagnostic accuracy in detecting tumor-specific variation from DNA extracted from samples collected through minimally invasive procedures (4-7). In addition to plasma samples, malignant non-blood biological fluids in close contact with the tumors, including pleural effusion, ascites, and cerebrospinal fluid, and cytological specimens are now widely used as specimens for NGS applications (8-12). Other easily accessible biological fluids, including sputum, that likely contain tumor-derived DNA are being actively explored for variation detection (8,13-19). Sputum analysis for lung cancer diagnosis was first described in 1958 and considered a well-established diagnostic technique (20,21). Sputum is the combination of saliva, phlegm, and mucus derived from the upper respiratory tract. Sputum can be obtained either via spontaneous production or through induction by inhalation of warm saline aerosol (20,21). Sputum has been used to detect genetic and epigenetic alterations in patients with various stages of lung cancer and in cancer-free chronic smokers who are at higher risk of developing lung cancer (8,13,15-18,22-31). These studies have consistently demonstrated that induced sputum samples contain cfDNA derived from the lungs and lower respiratory tract and are attractive candidate liquid biopsy media for lung cancer diagnosis (13,15-18,22-31). Studies exploring the concordance in somatic variations detected in tissue, blood, and sputum samples remain limited. In this study, we investigated the utility of induced sputum obtained from treatment-naïve patients with NSCLC as a medium for NGS-based somatic variation profiling. We present the following article in accordance with the MDAR and STROBE reporting checklists (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1297/rc).
Methods
Patient selection
Treatment-naïve patients diagnosed with locally-advanced to advanced stage NSCLCs from Xiangya Hospital, Central South University between October 2018 and June 2019 were included in this study. The study was approved by the institutional ethics board of Xiangya Hospital, Central South University (approval No. 201911306) and conducted in accordance with the Declaration of Helsinki (as revised in 2013). All patients provided written informed consent for the use of their biological samples.
Collection and preparation of sputum samples
The lung function of each patient was measured by spirometry as forced expiratory volume (FEV1), before and 10 minutes after, inhalation of 400 µg albuterol. Sputum induction was performed with hypertonic saline (4.5%) inhalation for 15 minutes for patients with FEV1 ≥1 L and isotonic saline (0.9%) for patients with impaired lung function (FEV1 <1 L). An aliquot of the expectorate was reserved for NGS and another for cytology analysis.
The induced sputum samples (~8 mL) were treated with 0.25% pancreatin at 37 ℃ with agitation at 660 rpm for 30 minutes. The digestion condition was adjusted to a maximum of 1:2.5 sputum to pancreatin ratio and/or extension of incubation time until complete liquefaction. The digestate was centrifuged (3,000 ×g, 10 minutes, 4 ℃); the supernatant was transferred to fresh tubes, centrifuged to remove cell debris (16,000 ×g, 10 minutes, 4 ℃), aliquoted into fresh tubes, and stored at −80 ℃ until DNA extraction. The remaining sediment was reconstituted in 1 mL supernatant and stored at −80 ℃ until DNA extraction.
DNA isolation and capture-based targeted NGS
The DNA isolation and targeted sequencing were performed at Burning Rock Biotech, a College of American Pathologists (CAP)-accredited/Clinical Laboratory Improvement Amendments (CLIA)-certified clinical laboratory, according to optimized protocols as described previously (9,10). Tissue DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) tissue biopsy samples and sputum-sediment samples using QIAamp DNA regular or FFPE tissue kits (Qiagen, Hilden, Germany). We extracted cfDNA from 4–5 mL of plasma samples, and 15 mL sputum-supernatant samples using a QIAamp Circulating Nucleic Acid kit (Qiagen, Germany). Target capture was performed using a commercial 168-gene panel. Indexed samples were sequenced on Nextseq500 (Illumina, Inc., San Diego, CA, USA) with paired-end reads and average sequencing depth of 1,000× for tumor samples and 10,000× for plasma, sputum supernatant, and sputum sediment samples. The NGS data analysis was performed using proprietary variant calling pipeline as described previously (9,10).
Statistical analysis
Variation detection rate was defined as the proportion of samples detected with variations relative to the total number of samples of the same sample type. Maximum allelic fraction (maxAF) was defined as the maximum fraction of the mutant allele detected from a sample, regardless of variation or gene. The concordance rate was defined as the proportion of the total number of variations detected from one sample type relative to the reference sample type. Statistical analyses were performed using the Fisher’s exact test, paired Student’s t-test, and Wilcoxon signed-rank test, as applicable, in R software (The R Foundation for Statistical Computing, Vienna, Austria). A P value <0.05 was considered statistically significant.
Results
Patient characteristics
This study included a total of 41 treatment-naïve patients with NSCLC, the baseline clinicopathologic features of whom are summarized in Table 1. Males comprised 68.3% (28/41) of the cohort, with a median age of 65 (range, 36 to 81 years). The majority had lung adenocarcinoma (78.0%; 32/41); the remaining patients had squamous cell carcinoma (n=7), and neuroendocrine tumor (n=2). Except for 3 patients with stage IIIA disease, the majority of patients (92.7%; 38/41) had stage IIIB-IV.
Table 1
| Clinicopathologic features | n=41 (%) |
|---|---|
| Age (years), median [range] | 65 [36–81] |
| Gender | |
| Male | 28 (68.3) |
| Female | 13 (21.7) |
| Smoking status | |
| Smoker | 27 (65.9) |
| Never smoker | 14 (34.1) |
| Histology | |
| Lung adenocarcinoma | 32 (78.0) |
| Lung squamous cell carcinoma | 7 (17.1) |
| NSCLC not otherwise specified | 2 (4.9) |
| Degree of cellular differentiation of sputum cytology | |
| Low | 20 (48.9) |
| Medium | 9 (22.0) |
| High | 7 (17.1) |
| NA | 5 (12.2) |
| Location of primary tumor | |
| Central | 22 (53.7) |
| Peripheral | 19 (46.3) |
| Stage | |
| ≤ IIIA | 3 (7.3) |
| IIIB–IIIC | 7 (17.1) |
| IV | 31 (75.6) |
NA, not applicable; NSCLC, non-small-cell lung cancer.
Sample distribution and quality control
All patients provided matched tumor, blood, and sputum samples; however, some of the samples were excluded due to insufficient volume (n=23) and inadequate DNA quality for library construction (n=2). Table S1 summarizes the samples available for matched analysis.
We extracted DNA from a total of 141 available samples, with an average DNA yield of 1,571.8 ng for tumor, 121.9 ng for plasma, 2,766.0 ng for sputum supernatant, and 8,144.5 ng for sputum sediment (Figure S1A). The tumor and sputum sediments samples had similar library complexity and insert size distribution and were distinct from plasma and sputum supernatant samples (Figure S1B). A majority of the tumor and sputum sediment samples had insert sizes of 150–250 base pairs; while plasma and sputum supernatant samples were smaller at 150–175 base pairs (Figure S1B). The median sequencing depth achieved 1,275× for tumor samples, 16,326× for plasma samples, 10,549× for sputum supernatant, and 16,660× for sputum sediment (Figure S1C).
We then compared the somatic variation detection rates and maxAF of plasma, sputum supernatant, and sputum sediment samples using matched tumor samples as reference. Relative to tumor samples, the somatic variation detection rates for the 168 genes were 76.9% for plasma (n=39), 72.4% for sputum supernatant (n=29), and 65.7% for sputum sediment samples (n=35) (Figure 1A). Meanwhile, the variation detection rates for the 8 classic NSCLC oncogenic driver genes and TP53 (9 genes) were 71.8% for plasma, 62.1% for sputum supernatant, and 51.4% for sputum sediment samples (Figure 1B). Using tumor samples as reference, the positive predictive value (PPV) was 80.4% for sputum supernatant, 55.6% for sputum sediment, and 73.3% for plasma samples. Meanwhile, the sensitivity was 36.9% for sputum supernatant, 31.3% for sputum sediment, and 50.0% for plasma, respectively, when considering the 168 genes. When considering only the nine genes, the PPV was 85.7% for sputum supernatant, 86.7% for sputum sediment, and 90.9% for plasma samples, while the sensitivity was 50.0% for sputum supernatant, 39.4% for sputum sediment, and 51.3% for plasma samples, respectively. As compared to tumor samples, the maxAF was significantly lower in plasma (P<0.001), sputum supernatant (P<0.001), and sputum sediment (P<0.001) in either the 168 genes or the 9 genes (Figure 1C,1D). However, maxAF was similar in plasma and sputum supernatant samples in either the 168 genes (P=0.81; Figure 1C) or the nine genes (P=0.55; Figure 1D).
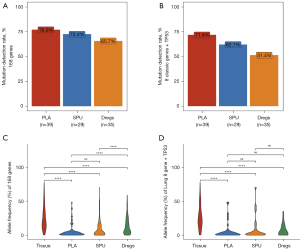
These data indicate that the DNA extracted from sputum supernatant and sputum sediment samples have adequate quality and sufficient quantity for NGS-based somatic variation profiling.
Somatic variation detection in sputum supernatant and sediment
Genomic profiling of sputum supernatant samples detected a total of 106 variations in 52 genes from 21 patients, with a detection rate of 72.4% (Figure 2A). The most frequent variations detected from sputum supernatants were TP53 (31.0%), EGFR (13.8%), KRAS (13.8%), and ALK (13.8%). Among the 8 genes, actionable variations were detected in 14 patients, including EGFR variations (p.L858R, n=1; p.E746_A750del n=3), EML4-ALK fusions (n=4), KRAS G12V/C/D/Q61H variations (n=4), ERBB2 A622S variation (n=1), and CD74-ROS1 fusion (n=1). No variations were detected in BRAF, MET, and RET from our cohort.
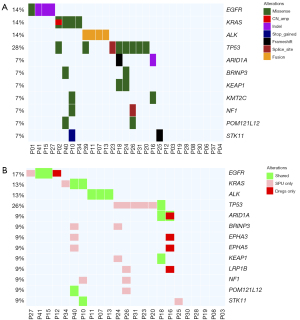
Meanwhile, a total of 276 variations in 75 genes were detected from matched sputum sediment samples from 14 patients, revealing a detection rate of 60.9%. Somatic variation detection rates were comparable between sputum supernatant and its corresponding sediment (78.3% vs. 65.2%; P=0.51). Considering the overall number of somatic variations detected from 23 patients with both sample types (Figure 2B), the variations detected from sputum supernatant and sediment samples were 47.8% concordant. Based on the distribution of variation types across the 168 genes, concordance rate was 80.0% for fusions and 30.4% for single nucleotide variants (SNVs) and short insertions/deletions (indels). A copy number variant (CNV) was only detected in 1 sputum supernatant sample and not detected in any sputum sediment sample (Table S2). Actionable variations were detected from the matched sputum supernatant and sediment of 8 patients, including EML4-ALK fusions (n=3), EGFR exon 19 deletion (19del) (n=2), KRAS G12C/Q61H mutations (n=2), and CD74-ROS1 fusion (n=1). Moreover, sputum supernatant samples had significantly higher maxAF than their corresponding sediment samples (P<0.001). The median maxAF was 1.26% (range, 0.0 to 9.2%) in sputum supernatant and 0.79% (range, 0.0 to 14.1%) in sputum sediment.
These data suggest the utility of DNA extracted from both sputum supernatant and sediment for NGS; however, the abundance of variations was significantly higher in sputum supernatant than its corresponding sediment.
Concordance of sputum supernatant and sediment with matched tumor sample
Figure 3A illustrates the somatic variation profile of tumor samples. Comparing the variation profile of 26 patients with both the sputum supernatant and tumor samples, 41 variations were detected from both samples (Figure 3B), with a concordance rate of 69.2%. The detection of fusions from both sputum supernatant and matched tumor samples was highly concordant, achieving 75.0%. The SNVs and indels were only 34.0% concordant, while CNVs were only detected from 2 sputum supernatant samples resulting in a 7.7% concordance (Table S3). It is worth noting that actionable fusions including EML4-ALK and CD74-ROS1 can be detected in sputum supernatant samples with a high concordance of 83.3% (5/6) relative to tumor samples (Table S3). Meanwhile, analysis of the variation profile of 32 patients with both sputum sediment and tumor samples demonstrated the detection of 45 variations from both samples (Figure 3C), with a concordance rate of 37.8%. The concordance rates relative to tumor samples of sputum supernatants were significantly higher than sputum sediments (69.2% vs. 37.8%; P=0.031).
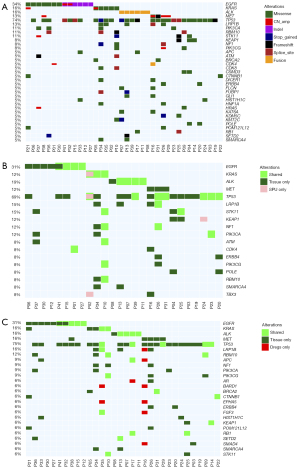
These data indicate that sputum supernatant samples are better than their sediment fraction in reflecting tumor-related variations and raise the need to fractionate the induced sputum samples for improving variation detection.
Concordance of sputum supernatant with matched plasma sample
Comparing the somatic variation profile of 28 patients with both sputum supernatant and plasma samples (Figure 4A), 32 variations were detected from both samples, with a concordance rate of 53.6% (Figure 4B). Sputum supernatant and plasma samples had comparable detection rates (71.4% vs. 67.9%; P=1.00) but significantly higher median allelic fraction than their matched plasma samples (P=0.034). Sputum supernatant and matched plasma samples were highly concordant in detecting fusions, achieving 83.5%. The SNVs and indels were 31.0% concordant, while CNVs were only detected from 3 sputum supernatant samples, resulting in a 7.7% concordance (Table S4).
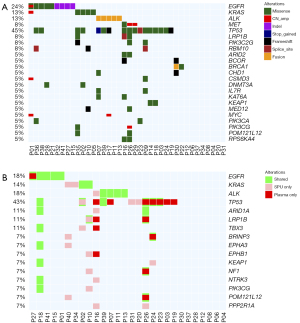
These data indicate the comparable somatic variation detection rates between sputum supernatant and plasma samples, suggesting the utility of sputum supernatant as an alternative sample for somatic variation profiling, particularly for non-CNV variations.
Sputum supernatant from smokers and non-smokers
Next, we investigated the clinical factors that are associated with better variation detection for induced sputum samples. All the clinical features analyzed, including age, gender, disease stage, smoking history, and histology, were not statistically correlated with variation detection rate in either sputum supernatant or sediment samples (Table S5). However, significantly higher maxAF (P=0.018; Table S5) and AF (P=0.021; Figure S2) were observed in the sputum supernatant samples from smokers than from non-smokers, suggesting that sputum supernatant samples, particularly from smokers, could provide valuable genetic information.
Case vignette
Of the 13 patients evaluable for sputum cytology, 38.5% (5/13) were identified with malignant cells. Figure 5 illustrates the apparent heterogeneous cell nuclei in the sputum cytology of samples from 3 patients diagnosed with stage IVA–IVB lung cancer of various histologies.
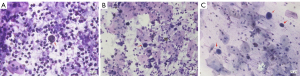
Figure 5A displays the sputum cytology findings for patient P23, a 56-year-old female non-smoker diagnosed with stage IVA pulmonary sarcomatoid carcinoma. The variations TP53 c.993+1G>C and PIK3CA p.H1047R were detected from both the matched tissue and sputum supernatant samples but were undetected from the sputum sediment sample. With no actionable variations detected, she received pemetrexed, carboplatin, and bevacizumab as front-line therapy.
Figure 5B shows the sputum cytology findings for patient P27, a 55-year-old male smoker diagnosed with stage IVB well-differentiated squamous cell lung carcinoma. The EGFR 19del E746_A750 was detected from his matched tissue, plasma, and sputum supernatant samples, but was undetected from the sputum sediment sample. His disease achieved partial response with cisplatin, paclitaxel, and pembrolizumab as the front-line regimen. Upon detection of EGFR 19del, he received icotinib as the second-line regimen.
Figure 5C shows the sputum cytology findings for patient P41, a 66-year-old female non-smoker diagnosed with stage IVB poorly-differentiated lung adenocarcinoma. The EGFR 19del was detected from all her samples, including tissue, plasma, sputum supernatant, and sediment samples. In addition to the EGFR 19del, EGFR copy number amplification, and TP53 c.783-1G>T were detected from her tumor sample, which were undetected in other sample types. She received icotinib as the front-line regimen and achieved complete response.
Discussion
Exfoliative cytology, which involves the microscopic study of the cells exfoliated from tumors in various samples, including saliva, sputum, and bronchial secretions, has been well-established as a non-invasive procedure in providing diagnostic information (32,33). Numerous studies have also established that cfDNA derived from the lower respiratory tract can be extracted from various sample types such as bronchoalveolar lavage fluid (BALF) and induced sputum (13-19,22,29,34). The collection of induced sputum involves inhalation of warm saline aerosol produced from an ultrasonic nebulizer and is non-invasive. Contrastingly, BALF is minimally invasive and is collected using bronchoalveolar lavage, which involves the insertion of a flexible bronchoscope thru the mouth or nose to reach the lungs and typically requires sedation (35). Due to accessibility and safety, induced sputum is a more attractive specimen type than BALF. The diagnostic accuracy of sputum cytology for lung cancer diagnosis has been demonstrated to achieve a specificity of 90%, sensitivity of 87%, and PPV of 79%, which in the absence of necrotizing pneumonia could exceed 95% (32). Sputum samples have been explored in polymerase chain reaction (PCR)-based detection of gene variations (13,14,16,18,22,29); however, only limited studies have reported their utility in NGS-based genomic profiling (15,17,34). In our study, we demonstrated the feasibility of using sputum supernatant as an alternative liquid biopsy specimen for NGS-based somatic variation profiling. The quality and quantity of cfDNA isolated from the supernatant fraction of induced sputum samples were adequate for NGS-based applications. Based on its significantly higher PPV (80.4% vs. 55.6%) and concordance with tumor samples (69.2% vs. 37.8%; P=0.031), sputum supernatant is the optimal fraction for the accurate detection of tumor-related non-CNV variations than its sediment fraction, raising the need to fractionate the induced sputum samples. The higher concordance with tumor samples also raises 3 important points: First, the sediment fraction might be comprised of a majority of cell debris and their removal does not hinder the detection of tumor-related variations from the supernatant fraction. Second, the concentration of circulating tumor DNA (ctDNA) found in sputum supernatant is higher than the sediment fraction, implying that the use of sputum without fractionation could dilute the sample and risk missing the detection of some clinically relevant variations. Third, the molecular profile derived from sputum supernatant samples more accurately reflects the non-CNV variations found in the primary lung tumor, which is more clinically relevant. The overall concordance in variations detected from sputum samples relative to the matched tumor samples observed from our cohort (69.2%) was consistent with the overall concordance of 74% reported by Wu et al. (15). The comparable variation detection rates, particularly in actionable non-CNV variations, between sputum supernatant and plasma samples further suggest the feasibility of using sputum as an alternative liquid biopsy specimen. Similar to the observations by Wu et al. (15), our study also demonstrated differences and similarities in variation profiles in matched sputum fractions, plasma, and tumor tissues, which might be related to spatial genetic heterogeneity inherent in small volume needle biopsy samples.
In clinical practice, induced sputum can serve as an alternative to minimize the need for obtaining tumor samples using tissue biopsy or when tumor or blood samples are difficult to obtain. Previous studies have demonstrated the feasibility of detecting genetic and epigenetic alterations in sputum samples from cancer-free chronic smokers as a strategy for early detection of lung cancer (13,22,23,26,29). The NGS-based somatic variation profiling of sputum supernatant samples was adequately sensitive in detecting actionable variations, particularly fusions, which has clinical value in guiding the use of appropriate targeted therapies. A limitation of using sputum samples in clinical diagnosis is its low sensitivity in detecting CNVs, which is due to the smaller fragments of cfDNA present in the sputum and limits the detection of CNVs from cfDNA in general.
Our study was limited by being conducted in a single-center and the small cohort size. A study with a larger cohort is warranted.
Conclusions
Our study demonstrates that the supernatant fraction of induced sputum from patients with advanced-stage NSCLC could serve as an alternative source of tumor DNA for comprehensive somatic variation profiling. Our study contributes to the growing number of sample types, besides plasma and tissue samples, that can provide accurate genomic data, particularly actionable non-CNV variations, for guiding therapeutic decisions in patients with lung cancer.
Acknowledgments
The authors would like to thank all the patients and their family members for their cooperation and support. We also thank the investigators, study coordinators, operation staff, and the whole project team who worked on this project.
Funding: This study was supported by grants from the Hunan Provincial Medical Technology Innovation Guidance Program (No. 2020SK53703 to LQ) and the General Program of the Natural Science Foundation of Hunan Province (No. 2021JJ31090 to LQ). The funders had no role in the conceptualization, design, data collection, analysis, decision to publish, or preparation of the manuscript.
Footnote
Reporting Checklist: The authors have completed the MDAR and STROBE reporting checklists. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1297/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1297/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1297/coif). AL, BL, SZ, and ZC are employed by Burning Rock Biotech. LQ received funding from the Hunan Provincial Medical Technology Innovation Guidance Program (No. 2020SK53703) and the General Program of the Natural Science Foundation of Hunan Province (No. 2021JJ31090). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the institutional ethics board of Xiangya Hospital, Central South University (approval No. 201911306) and informed consent was taken from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Colomer R, Mondejar R, Romero-Laorden N, et al. When should we order a next generation sequencing test in a patient with cancer? EClinicalMedicine 2020;25:100487. [Crossref] [PubMed]
- Mosele F, Remon J, Mateo J, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol 2020;31:1491-505. [Crossref] [PubMed]
- Peng M, Chen C, Hulbert A, et al. Non-blood circulating tumor DNA detection in cancer. Oncotarget 2017;8:69162-73. [Crossref] [PubMed]
- Rossi G, Ignatiadis M. Promises and Pitfalls of Using Liquid Biopsy for Precision Medicine. Cancer Res 2019;79:2798-804. [Crossref] [PubMed]
- Kim L, Tsao MS. Tumour tissue sampling for lung cancer management in the era of personalised therapy: what is good enough for molecular testing? Eur Respir J 2014;44:1011-22. [Crossref] [PubMed]
- Balla A, Hampel KJ, Sharma MK, et al. Comprehensive Validation of Cytology Specimens for Next-Generation Sequencing and Clinical Practice Experience. J Mol Diagn 2018;20:812-21. [Crossref] [PubMed]
- Heitzer E, Haque IS, Roberts CES, et al. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet 2019;20:71-88. [Crossref] [PubMed]
- Takano T, Ohe Y, Tsuta K, et al. Epidermal growth factor receptor mutation detection using high-resolution melting analysis predicts outcomes in patients with advanced non small cell lung cancer treated with gefitinib. Clin Cancer Res 2007;13:5385-90. [Crossref] [PubMed]
- Mao X, Zhang Z, Zheng X, et al. Capture-Based Targeted Ultradeep Sequencing in Paired Tissue and Plasma Samples Demonstrates Differential Subclonal ctDNA-Releasing Capability in Advanced Lung Cancer. J Thorac Oncol 2017;12:663-72. [Crossref] [PubMed]
- Li YS, Jiang BY, Yang JJ, et al. Unique genetic profiles from cerebrospinal fluid cell-free DNA in leptomeningeal metastases of EGFR-mutant non-small-cell lung cancer: a new medium of liquid biopsy. Ann Oncol 2018;29:945-52. [Crossref] [PubMed]
- Guo Z, Xie Z, Shi H, et al. Malignant pleural effusion supernatant is an alternative liquid biopsy specimen for comprehensive mutational profiling. Thorac Cancer 2019;10:823-31. [Crossref] [PubMed]
- Pisapia P, Pepe F, Iaccarino A, et al. Next Generation Sequencing in Cytopathology: Focus on Non-Small Cell Lung Cancer. Front Med (Lausanne) 2021;8:633923. [Crossref] [PubMed]
- Mao L, Hruban RH, Boyle JO, et al. Detection of oncogene mutations in sputum precedes diagnosis of lung cancer. Cancer Res 1994;54:1634-7. [PubMed]
- Boldrini L, Gisfredi S, Ursino S, et al. Mutational analysis in cytological specimens of advanced lung adenocarcinoma: a sensitive method for molecular diagnosis. J Thorac Oncol 2007;2:1086-90. [Crossref] [PubMed]
- Wu Z, Yang Z, Li CS, et al. Differences in the genomic profiles of cell-free DNA between plasma, sputum, urine, and tumor tissue in advanced NSCLC. Cancer Med 2019;8:910-9. [Crossref] [PubMed]
- Wang Z, Zhang L, Li L, et al. Sputum Cell-Free DNA: Valued Surrogate Sample for Detection of EGFR Mutation in Patients with Advanced Lung Adenocarcinoma. J Mol Diagn 2020;22:934-42. [Crossref] [PubMed]
- Nishikawa T, Fujii T, Tatsumi S, et al. Molecular Analysis of Liquid-Based Cytological Specimen Using Virtually Positive Sputum with Adenocarcinoma Cells. Diagnostics (Basel) 2020;10:84. [Crossref] [PubMed]
- Isaka T, Yokose T, Ito H, et al. Detection of EGFR mutation of pulmonary adenocarcinoma in sputum using droplet digital PCR. BMC Pulm Med 2021;21:100. [Crossref] [PubMed]
- Zeng D, Wang C, Mu C, et al. Cell-free DNA from bronchoalveolar lavage fluid (BALF): a new liquid biopsy medium for identifying lung cancer. Ann Transl Med 2021;9:1080. [Crossref] [PubMed]
- Bickerman HA, Sproul EE, Barach AL. An aerosol method of producing bronchial secretions in human subjects: a clinical technic for the detection of lung cancer. Dis Chest 1958;33:347-62. [Crossref] [PubMed]
- Weiszhar Z, Horvath I. Induced sputum analysis: step by step. Breathe 2013;9:300-6. [Crossref]
- Kersting M, Friedl C, Kraus A, et al. Differential frequencies of p16(INK4a) promoter hypermethylation, p53 mutation, and K-ras mutation in exfoliative material mark the development of lung cancer in symptomatic chronic smokers. J Clin Oncol 2000;18:3221-9. [Crossref] [PubMed]
- Belinsky SA, Klinge DM, Dekker JD, et al. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin Cancer Res 2005;11:6505-11. [Crossref] [PubMed]
- Belinsky SA, Liechty KC, Gentry FD, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res 2006;66:3338-44. [Crossref] [PubMed]
- Machida EO, Brock MV, Hooker CM, et al. Hypermethylation of ASC/TMS1 is a sputum marker for late-stage lung cancer. Cancer Res 2006;66:6210-8. [Crossref] [PubMed]
- Castagnaro A, Marangio E, Verduri A, et al. Microsatellite analysis of induced sputum DNA in patients with lung cancer in heavy smokers and in healthy subjects. Exp Lung Res 2007;33:289-301. [Crossref] [PubMed]
- Li R, Todd NW, Qiu Q, et al. Genetic deletions in sputum as diagnostic markers for early detection of stage I non-small cell lung cancer. Clin Cancer Res 2007;13:482-7. [Crossref] [PubMed]
- van der Drift MA, Prinsen CF, Hol BE, et al. Can free DNA be detected in sputum of lung cancer patients? Lung Cancer 2008;61:385-90. [Crossref] [PubMed]
- Baryshnikova E, Destro A, Infante MV, et al. Molecular alterations in spontaneous sputum of cancer-free heavy smokers: results from a large screening program. Clin Cancer Res 2008;14:1913-9. [Crossref] [PubMed]
- Katz RL, Zaidi TM, Fernandez RL, et al. Automated detection of genetic abnormalities combined with cytology in sputum is a sensitive predictor of lung cancer. Mod Pathol 2008;21:950-60. [Crossref] [PubMed]
- Hulbert A, Jusue-Torres I, Stark A, et al. Early Detection of Lung Cancer Using DNA Promoter Hypermethylation in Plasma and Sputum. Clin Cancer Res 2017;23:1998-2005. [Crossref] [PubMed]
- Jay SJ, Wehr K, Nicholson DP, et al. Diagnostic sensitivity and specificity of pulmonary cytology: comparison of techniques used in conjunction with flexible fiber optic bronchoscopy. Acta Cytol 1980;24:304-12. [PubMed]
- Hubers AJ, Prinsen CF, Sozzi G, et al. Molecular sputum analysis for the diagnosis of lung cancer. Br J Cancer 2013;109:530-7. [Crossref] [PubMed]
- Wu Z, Yang Z, Li CS, et al. Non-invasive detection of EGFR and TP53 mutations through the combination of plasma, urine and sputum in advanced non-small cell lung cancer. Oncol Lett 2019;18:3581-90. [Crossref] [PubMed]
- Davidson KR, Ha DM, Schwarz MI, et al. Bronchoalveolar lavage as a diagnostic procedure: a review of known cellular and molecular findings in various lung diseases. J Thorac Dis 2020;12:4991-5019. [Crossref] [PubMed]
(English Language Editor: J. Jones)


