
Molecular characterization of vascular intestinal obstruction using whole-exome sequencing
Introduction
Intestinal obstruction is a pathology commonly encountered in emergency and surgical departments, which is defined by the cessation of bowel function whatever the cause (1). Intra-abdominal adhesions, malignancy, or intestinal herniation often result in acute intestinal obstruction which interrupts the flow of intestinal contents (2). The main clinical presentation includes nausea and vomiting, abdominal distension, abdominal pain, and anal exhaust stop (3). Laboratory evaluation should include a complete blood count and metabolic panel (3). Radiography or computed tomography can accurately diagnose intestinal obstruction and assist in developing treatment strategies (2). Management of intestinal obstruction includes fluid resuscitation with correction of physiologic derangements caused by the obstruction, intestinal decompression, and bowel rest (3). With the advent of more sophisticated diagnostic tests, the morbidity and mortality associated with acute intestinal obstruction have declined, but its clinical management remains challenging (4). Intestinal obstruction can be divided into simple obstruction and strangulated intestinal obstruction. Acute vascular intestinal obstruction belongs to the category of strangulated intestinal obstruction, and has a rapid progression, poor prognosis, and high mortality (5).
Next-generation sequencing (NGS) is transforming clinical research and diagnostics, revolutionizing the field of genetic diagnosis and vastly enhancing our ability to perform comprehensive diagnostic testing in the clinic (6). Whole-exome sequencing (WES) is a widely used sequencing technology that can capture the majority of coding regions of the genome for sequencing, which contain the majority of disease-causing mutations, including point mutations (missense and nonsense mutations), small deletions and insertions in addition to canonical splicing mutations (7). It has emerged as a promising technique for searching for underlying genetic variations in various diseases, and revolutionized molecular diagnosis by providing an extraordinary opportunity to promptly identify genetic variations, including intestinal diseases (8). Mathur et al. identified rare variants linked to congenital pouch colon by performing WES (9). Chang et al. examined the mutational landscape of colorectal cancer in a Taiwanese population (10). Dong et al. identified a heterozygous frameshift myosin heavy chain 11 (MYH11) variant c.5819delC (p.Pro1940HisfsTer91) in chronic intestinal pseudo-obstruction (11). However, to the best of our knowledge, no previous study has explored the mutational landscape of vascular intestinal obstruction.
We conducted a study by WES of 9 sporadic patients with acute vascular intestinal obstruction. The mutation genes in each patient and the mutation genes shared by all 9 patients were identified. Next, a functional annotation analysis and protein-protein interaction (PPI) analysis of the shared mutation genes in the 9 patients were performed. Our study is the first to report the mutational landscape of sporadic vascular intestinal obstruction. We sought to detect several mutations associated with vascular intestinal obstruction, in the hope of providing a theoretical basis for the pathogenesis, diagnosis and treatment of vascular intestinal obstruction, so as to improve the diagnosis and treatment of vascular intestinal obstruction. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1134/rc).
Methods
Study subjects
The study cohort comprised 9 sporadic patients (5 females and 4 males), aged from 45 to 85 years, with acute vascular intestinal obstruction. None of the participants had a family history of vascular intestinal obstruction. Peripheral blood was collected from each participant for deoxyribonucleic acid (DNA) extraction. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethical committee of The First Affiliated Hospital of Bengbu Medical College (No. 2020KY077) and informed consent was taken from all individual participants.
WES and data analysis
The DNA samples were sequenced at the Beijing Genomics Institute, and successive data analyses were conducted. The raw data were mapped and aligned to the human genome GRCh37 (hg19) with Burrows-Wheeler Aligner-Maximal Exact Match. The duplicates were marked and removed by Picard tools. Local realignment was carried out with the Genome Analysis Toolkit (GATK) using different options, including Realigner Target Creator and Base Recalibrator, on the sorted binary alignment map files. The average read length was 150 bp. Single nucleotide polymorphisms (SNPs) and small indels were detected by the HaplotypeCaller module using the GATK software. ANNOVAR software was used to annotate the mutation sites. The mutations were annotated with genes, dbSNP ID, mutation types, mutation details, and mutation frequencies in the 1000 Genome and ExAC database. The functional effects of all the mutations on diseases were evaluated using the SIFT, PloyPhen, ClinVar, and InterVar databases. Next, mutations with mutation frequencies <0.01 in the 1000 Genome and EXAC databases, synonymous mutations and mutations sites located in introns, and harmless mutations in the SIFT, PloyPhen, ClinVar and InterVar databases were removed.
Functional annotation and PPI analysis of mutation genes
A Gene Ontology (GO) analysis was performed to explore the biological function of the identified mutation genes with the Database for Annotation, Visualization and Integrated Discovery (DAVID). The pathway annotation was conducted with the Reactome database. Additionally, String 11.0 was used to perform the PPI analysis on the proteins encoded by the mutated genes.
Copy number variation (CNV) identification
CNVs were identified using an open-source software called CNV Kit, a tool kit for inferring and visualizing copy number from targeted DNA-sequencing data, which takes advantage of both on- and off-target sequencing reads and applies a series of corrections to improve the accuracy of copy number calling (12).
Validation of candidate variants with quantitative real-time polymerase chain reaction (qRT-PCR)
In total, 23 blood samples were obtained from 13 patients with vascular intestinal obstruction and 10 healthy control patients. The total ribonucleic acid (RNA) was isolated with the Trizol reagent (Invitrogen, USA) in accordance with the manufacturer’s instructions. The qRT-PCR reactions were performed using the ABI 7300 Real-Time PCR Detection System with SuperReal PreMix Plus (Invitrogen, USA). Relative gene expression was analyzed using the 2−ΔΔCT method. The human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and actin beta (ACTB) were used as endogenous controls.
Statistical analysis
All of the statistical analyses were performed in R environment. In addition, the GraphPad Prism Software, version 7.0 was used for the statistical analysis of experimental data. The results are expressed as means ± standard deviations (SDs). One-way analysis of variance (ANOVA) was used to analyze mRNA expression. P<0.05 was considered to indicate a significant difference.
Results
Genetic analysis
On average, 159,542,590 clean reads were obtained per sample. The analysis identified 1,612 mutations in 600 genes in patient 1, 1,661 mutations in 663 genes in patient 2, 1,557 mutations in 623 genes in patient 3, 1,602 mutations in 681 genes in patient 4, 1,755 mutations in 730 genes in patient 5, 1,674 mutations in 667 genes in patient 6, 1,742 mutations in 697 genes in patient 7, 1,459 mutations in 641 genes in patient 8, 1,815 mutations in 690 genes in patient 9. By overlapping these genes with Venny, 112 mutation genes were identified as being shared by all 9 patients.
Functional annotation analysis and PPI analysis
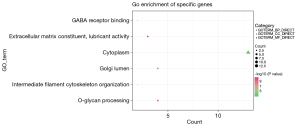
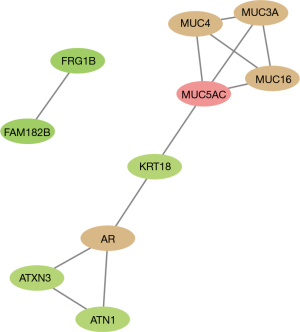
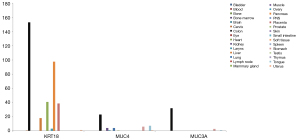
The GO analysis showed that the 112 mutated genes were significantly enriched in extracellular matrix constituent lubricant activity (P=2.90E-05), O-glycan processing (P=9.43E-05) and Golgi lumen (P=3.37E-04) (see Figure 1). The Reactome database revealed 2 significantly enriched pathways; that is, the O-linked glycosylation of the mucins (MUC) (P=4.52E-06) and the termination of O-glycan biosynthesis (P=5.51E-05). Notably, 4 genes (i.e., MUC16, MUC3A, MUC4, and MUC5AC) were enriched in the O-glycan processing, the O-linked glycosylation of the MUCs, and the termination of O-glycan biosynthesis. Additionally, a PPI network containing 10 proteins was established. Among them, MUC5AC was the protein with the highest degree (see Figure 2). The tissue differential expression of the above genes in normal individuals was obtained after a search of the Tissue-specific Gene Expression and Regulation (TiGER) database. Among them, keratin 18 (KRT18), MUC4, and MUC3A were significantly more highly expressed in the colon tissues than the other tissues (see Figure 3). Thus, we speculated that the genes encoding the MUC family were closely related to the occurrence of intestinal obstruction, among which the mutated genes in patients were MUC5AC, MUC16, MUC3A, and MUC4.
CNV identification
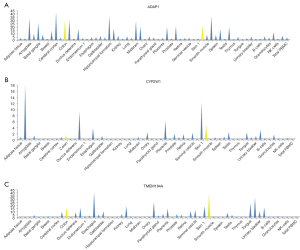
Using a CNV kit, chr7 13744_4856142 was found to be a deletion in 8 of the 9 patients. After deleting duplicates, a total of 17 genes were annotated within the Reference Sequence (ref-seq) database. Next, the tissue differential expressions of these 17 genes in normal individuals were queried using The Human Protein Atlas (https://www.proteinatlas.org/) database. Among them, ADAP1, CYP2W1, and TMEM184A were highly expressed in colon tissues and small intestine tissues (see Figure 4).
Validation of candidate variants
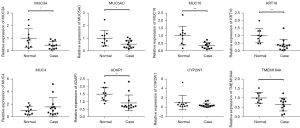
The expression levels of the 8 candidate genes between the vascular intestinal obstruction and normal control groups were measured with qRT-PCR. As Figure 5 shows, MUC5AC, MUC16, MUC3A, KRT18, ADAP1, and TMEM184A were significantly downregulated in the vascular intestinal obstruction group. No significant differences in the expression levels of MUC4 and CYP2W1 were observed between the vascular intestinal obstruction and healthy control groups.
Discussion
MUC proteins are highly glycosylated and belong to the family of large glycoproteins that constitute the major structural component of mucus (13). According to structural differentiation, MUCs can be classified into the following 2 categories: (I) secreted MUCs; and (II) membrane-bounded (transmembrane) MUCs. MUCs are found in all organs in contact with the external environment, including the gastrointestinal tract, respiration, reproductive tract, and ocular surface (13). A previous case report indicated that MUC may be a cause of early adhesional intestinal obstruction (14). In this study, we found that the MUC family members of MUC3A, MUC5AC and MUC16 were mutated in all patients with vascular intestinal obstruction.
MUC 3A is an intestinal membrane bound MUC encoded by the MUC3A gene residing at chromosome locus 7q22.1, while MUC5AC is a secreted gel forming MUC encoded by the gene MUC5AC that resides in the chromosome locus 11p15.5 (13). MUC3A has been found to be aberrantly expressed in various cancers, including pancreatic, gastric, renal, and breast cancer (13). Zhu et al. found that MUC3A was missense-mutated in each primary immune thrombocytopenia patient (n=20) (15). Salas et al. suggested that MUC3A carried significantly more pathogenic variations in patients with complicated cases of pneumococcal pneumonia (empyema) than control patients (16). Ren et al. reported that the variant c.1151T>C of the MUC3A gene may be a cause of intestinal symptoms in a Chinese pedigree with ankylosing spondylitis (17). Similarly, high MUC5AC expression has been frequently detected in multiple diseases. For example, the messenger RNA expression of MUC5AC was found to be enhanced in biliary tract cancer (18). The overexpression of MUC5AC was detected in colon cancer progression (19). Olli et al. indicated that MUC5AC expression protected the colonic barrier in experimental colitis (20). Our study identified 2 missense mutations in MUC3A (i.e., c.1033A>T:p.T345S and c.1091A>T:p.E364V) and a missense mutation in MUC5AC (i.e., g.1213256C>T:p.P4629L) in patients with vascular intestinal obstruction.
MUC16, which is encoded by the ~179 kb gene present on the short arm of human chromosome 19 at 19p13.2, is a membrane-tethered MUC protein with the following 3 components: (I) a C-terminal domain; (II) a tandem-repeat region; and (III) an extracellular N-terminal section (21). MUC16 has been extensively used as a biomarker for ovarian cancer, and its aberrant expression has been associated with several human malignancies, including pancreatic, breast, and lung cancer (22). Additionally, the MUC16 mutation has often been found in cancer cells. Li et al. reported that the MUC16 mutation was associated with a higher tumor mutation load and better survival outcomes in patients with gastric cancer (23). MUC16 has been found to be frequently mutated in hepatocellular carcinoma (24). Hu et al. demonstrated that MUC16 mutations were associated with the favorable prognosis of endometrial cancer patients (25). Ge et al. constructed a novel 5-gene (SMAD4, MUC16, COL6A3, FLG, and LRP1B) prognostic mutational signature in patients with stage III colon cancer (26). In the present study, we found 2 novel frameshift mutations in MUC16; that is, c.40673_40676del:p.K13558Tfs*13 and c.40672_40673insTCGG:p.K13558Ifs*24. Interestingly, the MUC3A, MUC5AC, and MUC16 proteins interacted with each other in the PPI network. This suggests that MUC3A, MUC5AC, and MUC16 interact harmoniously during vascular intestinal obstruction progression.
The type I intermediate filament chain KRT18 (or K18) encoded by KRT18 is located on chromosome 12, while all other type I keratins (except for K8) are located on the long arm of chromosome 17 (27). KRT18 and its filament partner K8 are perhaps the most commonly found members of the intermediate filament gene family. The high expression of KER18 has been found to be associated with a higher degree of malignancy and a worse prognosis for colorectal cancer patients (28). A review concluded that if mutations in these keratins cause human pathologies, they are likely to involve the liver or intestine (27). Mutations in KRT18 have been linked to cryptogenic cirrhosis (29). The K18 mutation (K18 S230T) was detected in a patient with inflammatory bowel disease (30). In this study, 3 novel missense mutations (i.e., c.127G>C:p.G43R, c.112G>T:p.G38C, and c.175G>T:p.G59W) were detected in vascular intestinal obstruction. Similarly, the KRT18 protein interacted with MUC5AC in the PPI network. Additionally, predictive software SIFT and PloyPhen showed that these mutations were harmful and destructive to protein function, which may indicate that the novel missense mutations in KRT18 may be a potential causative factor for vascular intestinal obstruction.
The orphan cytochrome P450 2W1 (CYP2W1) has been found to be exclusively expressed in transformed tissue in adult humans, mainly in colon tumors (31). High CYP2W1 levels have been observed in approximately 30% of human colorectal cancer specimens (32). Additionally, significantly elevated expression has also been observed in liver cancers with an even higher expression in liver metastases than in the parent tumor from the same patient (33). CYP2W1 has been reported to be related to the overall survival of left-sided colon cancer patients (34). In this study, CYP2W1 was found to be located in the copy number alteration deletion region in vascular intestinal obstruction, which may indicate a relationship between CYP2W1 and vascular intestinal obstruction.
In summary, we examined the mutational landscape of sporadic vascular intestinal obstruction for the first time. Apart from CYP2W1, multiple mutations in 4 genes (i.e., MUC3A, MUC5AC, MUC16, and KRT18) were identified in this study, which may be considered as candidate targets for the treatment of vascular intestinal obstruction. Our findings extend understandings of the potential pathological mechanism of vascular intestinal obstruction.
Acknowledgments
Funding: This work was funded by the Key Project of Natural Science Research Project of Anhui Provincial Education Department in 2021 (No. KJ2021A0767), and the Bengbu City-Level Scientific and Technological Innovation Guidance Project in 2021 (No. 20210332).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1134/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1134/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1134/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethical committee of The First Affiliated Hospital of Bengbu Medical College (No. 2020KY077) and informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hor T, Paye F. Diagnosis and treatment of an intestinal obstruction. Revue de l'infirmiere 2016;19-21. [Crossref] [PubMed]
- Jackson P, Vigiola Cruz M. Intestinal Obstruction: Evaluation and Management. Am Fam Physician 2018;98:362-7. [PubMed]
- Jackson PG, Raiji MT. Evaluation and management of intestinal obstruction. Am Fam Physician 2011;83:159-65. [PubMed]
- Margenthaler JA, Longo WE, Virgo KS, et al. Risk factors for adverse outcomes following surgery for small bowel obstruction. Ann Surg 2006;243:456-64. [Crossref] [PubMed]
- Min LI, Wu Y, Hua L, et al. Analysis of the characteristics of CT imaging of acute intestinal obstruction of blood circulation. China Modern Medicine 2014;21:124-5, 8.
- Mahajan MC, McLellan AS. Whole-Exome Sequencing (WES) for Illumina Short Read Sequencers Using Solution-Based Capture. Methods Mol Biol 2020;2076:85-108. [Crossref] [PubMed]
- Kryukov GV, Shpunt A, Stamatoyannopoulos JA, et al. Power of deep, all-exon resequencing for discovery of human trait genes. Proc Natl Acad Sci U S A 2009;106:3871-6. [Crossref] [PubMed]
- Simon AJ, Golan AC, Lev A, et al. Whole exome sequencing (WES) approach for diagnosing primary immunodeficiencies (PIDs) in a highly consanguineous community. Clin Immunol 2020;214:108376. [Crossref] [PubMed]
- Mathur P, Medicherla KM, Chaudhary S, et al. Whole exome sequencing reveals rare variants linked to congenital pouch colon. Sci Rep 2018;8:6646. [Crossref] [PubMed]
- Chang YS, Lee CC, Ke TW, et al. Molecular characterization of colorectal cancer using whole-exome sequencing in a Taiwanese population. Cancer Med 2019;8:3738-47. [Crossref] [PubMed]
- Dong W, Baldwin C, Choi J, et al. Identification of a dominant MYH11 causal variant in chronic intestinal pseudo-obstruction: Results of whole-exome sequencing. Clin Genet 2019;96:473-7. [Crossref] [PubMed]
- Talevich E, Shain AH, Botton T, et al. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol 2016;12:e1004873. [Crossref] [PubMed]
- Dhanisha SS, Guruvayoorappan C, Drishya S, et al. Mucins: Structural diversity, biosynthesis, its role in pathogenesis and as possible therapeutic targets. Crit Rev Oncol Hematol 2018;122:98-122. [Crossref] [PubMed]
- Ugare G, Osakwe G, Djunda E. Mucin as possible cause of early adhesional intestinal obstruction. Afr Health Sci 2014;14:954-8. [Crossref] [PubMed]
- Zhu JJ, Yuan D, Sun RJ, et al. Mucin mutations and aberrant expression are associated with the pathogenesis of immune thrombocytopenia. Thromb Res 2020;194:222-8. [Crossref] [PubMed]
- Salas A, Pardo-Seco J, Barral-Arca R, et al. Whole Exome Sequencing Identifies New Host Genomic Susceptibility Factors in Empyema Caused by Streptococcus pneumoniae in Children: A Pilot Study. Genes (Basel) 2018;9:240. [Crossref] [PubMed]
- Ren WF, Hu JT, Gao Y, et al. Whole exome sequencing in a pedigree with ankylosing spondylitis. Zhongguo Gu Shang 2020;33:672-6. [PubMed]
- Ruzzenente A, Iacono C, Conci S, et al. A novel serum marker for biliary tract cancer: diagnostic and prognostic values of quantitative evaluation of serum mucin 5AC (MUC5AC). Surgery 2014;155:633-9. [Crossref] [PubMed]
- Krishn SR, Kaur S, Smith LM, et al. Mucins and associated glycan signatures in colon adenoma-carcinoma sequence: Prospective pathological implication(s) for early diagnosis of colon cancer. Cancer Lett 2016;374:304-14. [Crossref] [PubMed]
- Olli KE, Rapp C, O'Connell L, et al. Muc5ac Expression Protects the Colonic Barrier in Experimental Colitis. Inflamm Bowel Dis 2020;26:1353-67. [Crossref] [PubMed]
- Haridas D, Ponnusamy MP, Chugh S, et al. MUC16: molecular analysis and its functional implications in benign and malignant conditions. FASEB J 2014;28:4183-99. [Crossref] [PubMed]
- Aithal A, Rauth S, Kshirsagar P, et al. MUC16 as a novel target for cancer therapy. Expert Opin Ther Targets 2018;22:675-86. [Crossref] [PubMed]
- Li X, Pasche B, Zhang W, et al. Association of MUC16 Mutation With Tumor Mutation Load and Outcomes in Patients With Gastric Cancer. JAMA Oncol 2018;4:1691-8. [Crossref] [PubMed]
- Zhan H, Jiang J, Sun Q, et al. Whole-Exome Sequencing-Based Mutational Profiling of Hepatitis B Virus-Related Early-Stage Hepatocellular Carcinoma. Gastroenterol Res Pract 2017;2017:2029315. [Crossref] [PubMed]
- Hu J, Sun J. MUC16 mutations improve patients' prognosis by enhancing the infiltration and antitumor immunity of cytotoxic T lymphocytes in the endometrial cancer microenvironment. Oncoimmunology 2018;7:e1487914. [Crossref] [PubMed]
- Ge W, Hu H, Cai W, et al. High-risk Stage III colon cancer patients identified by a novel five-gene mutational signature are characterized by upregulation of IL-23A and gut bacterial translocation of the tumor microenvironment. Int J Cancer 2020;146:2027-35. [Crossref] [PubMed]
- Owens DW, Lane EB. Keratin mutations and intestinal pathology. J Pathol 2004;204:377-85. [Crossref] [PubMed]
- Zhang J, Hu S, Li Y. KRT18 is correlated with the malignant status and acts as an oncogene in colorectal cancer. Biosci Rep 2019;39:BSR20190884. [Crossref] [PubMed]
- Ku NO, Darling JM, Krams SM, et al. Keratin 8 and 18 mutations are risk factors for developing liver disease of multiple etiologies. Proc Natl Acad Sci U S A 2003;100:6063-8. [Crossref] [PubMed]
- Zupancic T, Stojan J, Lane EB, et al. Intestinal cell barrier function in vitro is severely compromised by keratin 8 and 18 mutations identified in patients with inflammatory bowel disease. PLoS One 2014;9:e99398. [Crossref] [PubMed]
- Karlgren M, Gomez A, Stark K, et al. Tumor-specific expression of the novel cytochrome P450 enzyme, CYP2W1. Biochem Biophys Res Commun 2006;341:451-8. [Crossref] [PubMed]
- Choong E, Guo J, Persson A, et al. Developmental regulation and induction of cytochrome P450 2W1, an enzyme expressed in colon tumors. PLoS One 2015;10:e0122820. [Crossref] [PubMed]
- Stenstedt K, Hallstrom M, Lédel F, et al. The expression of CYP2W1 in colorectal primary tumors, corresponding lymph node metastases and liver metastases. Acta Oncol 2014;53:885-91. [Crossref] [PubMed]
- Liang L, Zeng JH, Qin XG, et al. Distinguishable Prognostic Signatures of Left- and Right-Sided Colon Cancer: a Study Based on Sequencing Data. Cell Physiol Biochem 2018;48:475-90. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


