
Identification of significant genes with a poor prognosis in skin cutaneous malignant melanoma based on a bioinformatics analysis
Introduction
Skin cutaneous malignant melanoma (SKCM) is a potentially fatal skin malignant tumor, and its incidence is increasing worldwide (1). At present, the average lifetime risk for melanoma has now reached 1 in 63 in the United States, and a similar probability has been reported in other Western countries (2). SKCM is rarer than other skin cancers; however, it is more lethal, and accounts for about 73% of skin cancer-related deaths (3). Early SKCM can be treated surgically, and the 5-year survival rate is as high as 90% (4). However, when patients are diagnosed with advanced or metastatic SKCM, the 5-year survival rate decreases significantly to 64% for regional malignant melanoma and 23% for distant malignant melanoma, and the options for treatment are very limited (5,6).
The emergence of immunotherapy and targeted therapy in recent years represents a breakthrough in the treatment of metastatic melanoma, and while immunotherapy has achieved certain results (7), its clinical application is limited due to its high immunotoxicity (8). Thus, modern molecular biology techniques and bioinformatics methods are being applied to study skin melanoma at the molecular level. The identification of the exact molecular mechanisms and metastasis-related biomarkers of cutaneous melanoma, and the identification of effective therapeutic targets, appear to be critical to the development of more effective therapies for cutaneous melanoma.
In this study, 3 data sets (GSE83583, GSE111766, and GSE104849) were chosen from Gene Expression Omnibus (GEO). In previous similar study (9), the GSE data sets were from different GPL platforms. It is well known that the gene annotation information of different GPL platforms is not the same, which ultimately leads to errors in research results. The GSE data sets in our study are all from the same GPL platform, which makes our final results more convincing. Next, the commonly differentially expressed genes (DEGs) in the 3 data sets were identified using the GEO2R online tool and R software with the ggplot2 and UpSet R packages. Next, the Database for Annotation, Visualization and Integrated DiscKery (DAVID) was used to test the DEGs to determine their molecular functions (MFs), cellular components (CCs), biological processes (BPs), and Kyoto Encyclopedia of Gene and Genome (KEGG) pathways. A protein-protein interaction (PPI) network was then formed, and the Cytotype Molecular Complex Detection (MCODE) plug-ins were used to further analyze and identify the core DEGs. The core DEGs were then imported into the software R with the survminer package to identify the significant prognostic information (P<0.05). Further, we validated the significantly expressed DEGs in the tumor stage I (T1), tumor stage II (T2), tumor stage III (T3) and tumor stage IV (T4) of melanoma stages via the R package, mainly ggplot2 (Version 3.3.3) (P<0.05). Notably, only 6 DEGs qualified for the clinical correlation analysis of tumor stage, which were subsequently re-analyzed via a KEGG pathway enrichment analysis. Ultimately, 2 DEGs [i.e., PRKCB and CYP1B1] were identified and found to be significantly enriched in the micro ribonucleic acids (RNAs) in the cancer pathway. The transcriptional levels were examined by quantitative real-time polymerase chain reaction, and the protein levels were obtained from the Human Protein Atlas (HPA). In conclusion, our bioinformatics study identified additional useful biomarkers that may serve as effective targets in the treatment of SKCM. We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1163/rc).
Methods
Microarray data information
The GEO of the National Center for Biotechnology Information is a public functional genomics data repository that contains 200,000+ human gene expression samples for data submissions (https://www.ncbi.nlm.nih.gov/geo/). We obtained the gene expression profiles of GSE83583, GSE111766, and GSE104849 in melanoma tissues and normal melanocyte tissues from this database. The microarray data of GSE83583, GSE111766, and GSE104849 all accessed from the GPL10558 Platform (Illumina HumanHT-12 V4.0 expression Bead chip). Among them, GSE83583 included 81 melanoma tissues and 3 normal melanocyte tissues, GSE111766 included 21 melanoma tissues and 3 normal melanocyte tissues, and GSE104849 included 22 melanoma tissues and 3 normal melanocyte tissues. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data filtering of DEGs
The DEGs of the melanoma and normal melanocyte specimens were identified using the GEO2R online tools (10). The filter criteria were as follows: a |logFC| >1.5 and an adjusted P value <0.05. The filtered data in TXT format were tested and visualized using R software (Version 3.5.3) by ggplot2 (Version 3.3.3) and UpSet R package (Version 1.4.0) to identify the common DEGs among the 3 data sets (11). The DEGs with a logFC <0 were considered downregulated genes, and those with a logFC >0 were considered upregulated genes.
Gene Ontology (GO) enrichment and KEGG pathway enrichment analyses
The GO enrichment analysis is a commonly used method to define genes and their protein or RNA products to identify the exclusive biological characteristics of high-throughput genomic data or transcriptomes (12). A KEGG analysis examines genomes, biological pathways, chemical materials, diseases, and drugs (13). The DAVID website (https://david.ncifcrf.gov/home.jsp) was designed to identify a large number of protein or gene functions (14). The enrichment of the DEGs in terms of their BPs, CCs, MFs, and KEGG pathways (P<0.05) was also analyzed by DAVID.
PPI network construction and module analysis
PPI information can be captured by the Search Tool for the Retrieval of Interacting Genes (STRING, https://cn.string-db.org/) (15). The STRING database (Version 11.5) and Cytoscape software (Version 3.6.0) (16) were used to check the potential physical and functional correlations among the DEGs (the conditions were a confidence score ≥0.4 and a maximum number of interactors =0). Further, the MCODE plug-in was used to check and visualize modules of the PPI network with the following parameter settings: node score cutoff =0.2; degree cutoff =2; maximum depth =100; and k-core =2.
Survival analysis, protein level, and RNA sequencing expression of core genes
The protein levels of the DEGs were examined by immunohistochemistry from the HPA(17) (http://www.proteinatlas.org/). R is a free software environment for statistical computing and graphics. We applied the R software for the statistical analysis and visualization of the data. The survival data and relevance (tumor stage) data were downloaded RNA Sequencing (RNAseq) data from the SKCM Project in the cancer genome atlas (TCGA) database (https://portal.gdc.cancer.gov/). Survminer R package (Version 0.4.9) (for the visualization of the Kaplan-Meier survival curves) and survival R package (Version 3.2–10) [for statistical the analysis of the survival data and clinical relevance (tumor stage) data] were used for the data analysis.
Statistical analysis
Statistical analysis was performed by R packages. All data in the study were presented as mean ± standard deviation (SD). Differences between two groups were determined using the Cox proportional hazards model. And the significance value was determined when P<0.05.
Results
Identification of DEGs in melanoma tissues
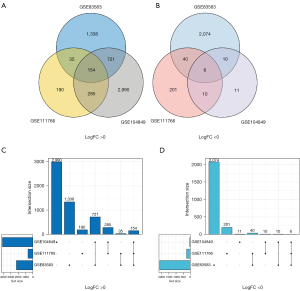
In the present study, the raw data of the 3 data sets contained a total of 124 melanoma tissues and 9 normal melanocyte tissues. GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=) was used to extract 4,378, 921, and 4,187 DEGs from the GSE83583, GSE111766, and GSE104849 data sets, respectively. All the 160 common DEGs were identified using R package (ggplot2 and UpSet). The DEGs included 154 upregulated genes (logFC >0) and 6 downregulated genes (logFC <0) in the melanoma tissues (see Figure 1 and Figure S1A).
GO and KEGG pathway analyses of DEGs in melanoma tissues
The GO annotation and KEGG pathway enrichment analyses of the 160 DEGs were conducted using the DAVID website. The results of the GO enrichment analysis were as follows: (I) in relation to the BPs, the upregulated DEGs were significantly enriched in the endothelial cell migration, positive regulation of endothelial cell proliferation, positive regulation of gene expression, cellular response to glucose stimulus, fatty acid metabolic process and signal transduction (no significant results were found for the downregulated DEGs); (II) in relation to the CCs, the upregulated DEGs were significantly enriched in oxidoreductase activity, peptide antigen binding, L-cystine transmembrane transporter activity, growth factor activity, and cytokine activity (no significant results were found for the downregulated DEGs); and (III) in relation to the MFs, the upregulated DEGs were significantly enriched in the extracellular exosome, extracellular space, lysosomal membrane, melanosome, endoplasmic reticulum membrane and lysosomal lumen (no significant results were found for the downregulated DEGs) (see Table 1).
Table 1
| Expression | Category | Term | Count | % | P value | FDR |
|---|---|---|---|---|---|---|
| Upregulated | GOTERM_BP_DIRECT | GO:0043542~endothelial cell migration | 4 | 2.649006623 | 0.001429913 | 1 |
| GOTERM_BP_DIRECT | GO:0001938~positive regulation of endothelial cell proliferation | 5 | 3.311258278 | 0.002010012 | 1 | |
| GOTERM_BP_DIRECT | GO:0010628~positive regulation of gene expression | 8 | 5.298013245 | 0.004314346 | 1 | |
| GOTERM_BP_DIRECT | GO:0006631~fatty acid metabolic process | 4 | 2.649006623 | 0.007600221 | 1 | |
| GOTERM_BP_DIRECT | GO:0071333~cellular response to glucose stimulus | 4 | 2.649006623 | 0.007600221 | 1 | |
| GOTERM_BP_DIRECT | GO:0007165~signal transduction | 18 | 11.9205298 | 0.008212234 | 1 | |
| GOTERM_CC_DIRECT | GO:0070062~extracellular exosome | 41 | 27.15231788 | 5.21E-05 | 0.009220898 | |
| GOTERM_CC_DIRECT | GO:0005615~extracellular space | 24 | 15.89403974 | 2.38E-04 | 0.016008507 | |
| GOTERM_CC_DIRECT | GO:0005765~lysosomal membrane | 10 | 6.622516556 | 2.71E-04 | 0.016008507 | |
| GOTERM_CC_DIRECT | GO:0042470~melanosome | 6 | 3.973509934 | 0.001092801 | 0.048356431 | |
| GOTERM_CC_DIRECT | GO:0005789~endoplasmic reticulum membrane | 16 | 10.59602649 | 0.002606126 | 0.092256876 | |
| GOTERM_CC_DIRECT | GO:0043202~lysosomal lumen | 5 | 3.311258278 | 0.004176494 | 0.123206574 | |
| GOTERM_MF_DIRECT | GO:0016491~oxidoreductase activity | 7 | 4.635761589 | 0.004053253 | 1 | |
| GOTERM_MF_DIRECT | GO:0042605~peptide antigen binding | 3 | 1.986754967 | 0.018681714 | 1 | |
| GOTERM_MF_DIRECT | GO:0015184~L-cystine transmembrane transporter activity | 2 | 1.324503311 | 0.029757723 | 1 | |
| GOTERM_MF_DIRECT | GO:0008083~growth factor activity | 5 | 3.311258278 | 0.034060443 | 1 | |
| GOTERM_MF_DIRECT | GO:0005125~cytokine activity | 5 | 3.311258278 | 0.043985806 | 1 | |
| Downregulated | None |
GO, Gene Ontology; DEG, differentially expressed gene; FDR, false discovery rate.
The KEGG analysis results are set out in Table 2. The upregulated DEGs were significantly enriched in Fc-gamma receptor-mediated phagocytosis, the erythroblastic oncogene B (erbB) signaling pathway, non-small cell lung cancer, and the prolactin signaling pathway, while the downregulated DEGs were not significantly enriched in any signaling pathways (P<0.05).
Table 2
| Pathway ID | Name | Count | % | P value | Genes |
|---|---|---|---|---|---|
| has04666 | Fc-gamma R-mediated phagocytosis | 5 | 3.31 | 0.005654767 | MARCKSL1, GSN, PRKCB, PIK3CD, AKT1 |
| hsa04012 | ErbB signaling pathway | 5 | 3.31 | 0.006400644 | STAT5A, ERBB3, PRKCB, PIK3CD, AKT1 |
| hsa05223 | Non-small cell lung cancer | 4 | 2.64 | 0.012041001 | PRKCB, PIK3CD, AKT1, FOXO3 |
| hsa04917 | Prolactin signaling pathway | 4 | 2.64 | 0.022686686 | STAT5A, PIK3CD, AKT1, FOXO3 |
KEGG, Kyoto Encyclopedia of Gene and Genome; DEG, differentially expressed gene.
PPI network construction and module analysis
In total, 160 DEGs were input into the DEG-PPI network complex to examine the molecular mechanisms regulating SKCM progression. The PPI interaction network included 210 edges and 89 nodes, and 88 upregulated genes were isolated, and 1 downregulated gene was isolated. Of the 160 DEGs, 71 were not contained in the DEG-PPI network (see Figure S1B1). Based on the PPI network, using Cytotype parameter settings, we applied the MCODE plug-in to further analyze the 89 nodes. The visual results identified 19 central nodes with upregulated genes (see Figure S1B2).
Analysis of core genes by the R software
The survminer package of R software was used to identify 19 core survival genes. Of these, 8 genes had a significantly worse overall survival in SKCM patient, while 11 had no significant effect (P<0.05; see Figure 2, Figure S1B3). Next, the R package, mainly ggplot2 (Version 3.3.3), was used to quantify the expression levels of the 8 genes between the cancerous and normal tissues. We found that CYP1B1, PRKCB, P450 cytochrome oxidoreductase (POR), CD79b molecule immunoglobulin-associated beta (CD79B), major facilitator superfamily domain containing 12 (MFSD12), and oculocutaneous albinism II (OCA2) had significant expression levels in the T1, T2, T3, and T4 SKCM tumor stages (P<0.05; see Figure 3 and Figure S1C).
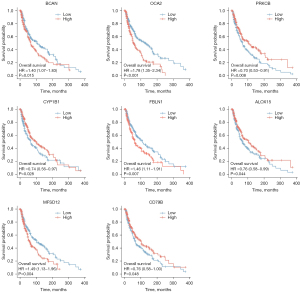
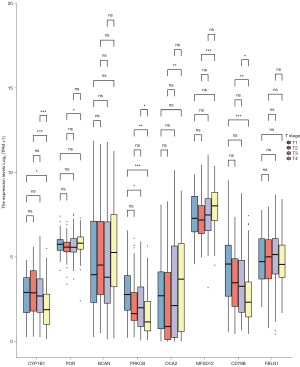
Re-analysis of the 6 high correlation genes via DAVID, and the effects of the protein levels of the core genes on the survival of patients
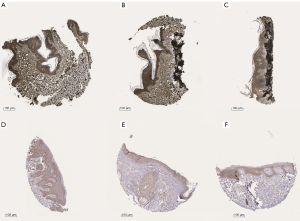
To interpret the potential pathways of these 6 genes, the KEGG pathway enrichment data were re-analyzed (P<0.05), and 2 genes (PRKCB and CYP1B1) were found to be significantly enriched in the microRNAs in the cancer pathway (see Table 3, Figure S1D, and Figure S2). The expression levels of PRKCB and CYP1B1 were detected using immunohistochemistry from the HPA. The protein levels of the 2 core genes were significantly higher in the SKCM tissues than the normal tissues (see Figure 4).
Table 3
| Pathway ID | Name | Count | % | Genes |
|---|---|---|---|---|
| hsa05206 | MicroRNAs in cancer | 2 | 33.33 | PRKCB, CYP1B1 |
KEGG, Kyoto Encyclopedia of Gene and Genome.
Discussion
In the current study, we used 3 profile data sets (GSE83583, GSE111766, and GSE104849) to identify useful prognostic biomarkers in SKCM using bioinformatics methods. The present research examined 124 melanoma specimens and 9 normal melanocytes specimens. By using the GEO2R and R software, 160 commonly changed DEGs, including 154 upregulated and 6 downregulated DEGs, were identified. The results of the GO and KEGG enrichment analyses were as follows: (I) in relation to the BPs, the upregulated DEGs were significantly enriched in the cellular response to glucose stimulus, signal transduction, positive regulation of gene expression, endothelial cell migration, fatty acid metabolic process, positive regulation of endothelial cell proliferation, extracellular exosome (no significant results were found for the downregulated DEGs); (II) in relation to the MFs, the upregulated DEGs were enriched in oxidoreductase, peptide antigen binding, L-cystine transmembrane transporter activity, growth factor activity, and cytokine activity (no significant results were found for the downregulated DEGs); and (III) in relation to the CCs, the upregulated DEGs were particularly enriched in the extracellular exosome, extracellular space, lysosomal membrane, melanosome, endoplasmic reticulum membrane, and lysosomal lumen (no significant results were found for the downregulated DEGs). The KEGG analysis revealed that the upregulated DEGs were significantly enriched in Fc-gamma receptor-mediated phagocytosis, the erbB signaling pathway, non-small cell lung cancer, the prolactin signaling pathway, while the downregulated DEGs were not enriched in any noteworthy pathways.
Using the STRING online database and Cytoscape software, we then constructed a DEG-PPI network complex, which included 89 nodes and 210 edges. The MCODE plug-in was used to screen 19 vital upregulated genes from the PPI network complex. Further, through the survminer package of the R software analysis and by plotting Kaplan-Meier survival curves, we discovered that 8 of the 19 genes had significantly worse overall survival in SKCM patients. In examining the 8 genes using R package (mainly ggplot2) we found that 6 genes were more highly expressed in the melanoma samples than the normal melanocyte samples. Ultimately, we re-analyzed 6 genes via a KEGG enrichment analysis and determined that 2 genes (i.e., PRKCB and CYP1B1) were enriched in the microRNAs of cancer tissues. These genes could potentially serve as novel molecular biomarkers and improve the diagnosis and prognosis of SKCM patients. In the future, we look forward to using computer AI-assisted technology and animal models to combine our experimental results to provide new candidate targets for the treatment of SKCM.
The CYP1B1 gene is a member of the extrahepatic xenobiotic-metabolizing enzyme family, which is known to encode 1 of the cytochrome P450 superfamily members of the enzyme. The cytochrome P450 proteins are monooxygenases that can accelerate the synthesis of steroids, other lipids, and cholesterol and drug metabolism. The enzyme encoded by this gene is located in the endoplasmic reticulum and metabolizes 17beta-estradiol, polycyclic aromatic hydrocarbons, and other carcinogens. A study of multiple biological pathways showed that single nucleotide polymorphisms (SNPs) near CYP1B1, which is a nevus gene, reached global significance in a bivariate analysis examining melanoma risk (18). Recent research has shown that certain germline mutations, such as CYP1B1, may contribute to the differential development of melanoma susceptibility (19). In 2016, Sumantran et al. reported that CYP1B1, which metabolizes most anti-cancer drugs, was more downregulated in melanoma samples than normal skin samples. The downregulation of CYP1B1 in melanoma prevents estrogen-dependent hormonal carcinogenesis while protecting the dermis and epidermis from activating many environmental mutagens (20). Additionally, the increased sensitivity to specific anti-cancer drugs led to the low expression of CYP1B1, and CYP1B1 depletion might have an anti-cancer effect on melanoma (20). Further, in the protein kinase A (PKA) and protein kinase C (PKC) pathways, the parathyroid hormone induces an increase in endothelial nitric oxide synthase activity (21). PKC beta II negatively regulates the HGF signaling process from growth factor receptor bound protein 2-associated protein 1 (Gab1) to phosphatidylinositol 3 kinase (PI3K), and the deletion of CYP1B1 is very important for melanoma cells to obtain invasive potential the demarcation of HLA-DR/CD18 complex within or outside lipid rafts (21).
PRKCB (also known as protein kinase C beta) is a serine- and threonine-specific protein kinase family that can be activated by second messenger diacylglycerol and calcium. PRKCB family members are known to be involved in a variety of cellular signaling pathways and act as major receptors for phorbol esters (tumor promoters) (22,23). A wide variety of protein targets are phosphorylated by PRKCB family members. The protein encoded by this gene is a member of the PRKCB family. This protein kinase has been reported to be involved in many different cellular functions, such as intestinal sugar absorption, apoptosis induction, endothelial cell proliferation, and B cell activation (24,25). In the 2018 World Health Organization’s “multidimensional” classification of cutaneous melanocytic neoplasms, it was pointed out that fusions studies of PRKCB and other genes have been used as advanced molecular studies to carefully distinguish between severely atypical (high-grade) melanocytomas and “classical” melanomas (26). Thus, the importance of PRKCA as a related gene in melanoma research is clear.
Previous study has shown that tyrosinase is a rate-limiting enzyme of melanin production that interacts with PRKCB and be activated (27). Additionally, the PRKCB mutation affects tyrosinase and its sequential signal transduction, which hinders melanin production and increases the risk of melanoma (28). Banerjee et al. (29) showed that the PRKCB mutation has a significant effect on 2 crucial serine residues at the C-terminal of tyrosinase protein. This mutation can inactivate tyrosinase, hinder melanogenesis, and strongly increase the risk of developing melanoma. Voris et al. (30) found that as the functions of PRKCB include oxidative stress response and melanin production, colony formation in soft agar is inhibited when PRKCB is re-expressed in melanoma cells; thus, the loss of PRKCB in melanoma plays an important role in melanoma growth.
Conclusions
This study used bioinformatics methods to identify 2 DEGs (i.e., CYP1B1and PRKCB) in melanoma and normal melanocyte tissues based on 3 different microarray data sets from the same GPL platform. The results showed that CYP1B1and PRKCB play critical roles in the process of SKCM development. However, further experiments need to be conducted to verify the function and mechanism prediction of these genes. These findings may provide insights into and guidance on the potential biomarkers, therapeutic targets, and biological mechanisms of SKCM.
Acknowledgments
Funding: This work was funded by the Tianjin Science and Technology Committee (grant No. 18YFZCSY01090).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1163/rc
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1163/coif). JW reports that this work was funded by the Tianjin Science and Technology Committee (grant No. 18YFZCSY01090). The other author has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Global Burden of Disease Cancer Collaboration. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol 2019;5:1749-68. [Crossref] [PubMed]
- Garbe C, Keim U, Gandini S, et al. Epidemiology of cutaneous melanoma and keratinocyte cancer in white populations 1943-2036. Eur J Cancer 2021;152:18-25. [Crossref] [PubMed]
- Shah R, Patel N, Patel Y, et al. Age Demographics of Subjects Enrolled in Global, Interventional Phase 3 Melanoma Clinical Trials. Ther Innov Regul Sci 2022;56:184-90. [Crossref] [PubMed]
- Moran B, Silva R, Perry AS, et al. Epigenetics of malignant melanoma. Semin Cancer Biol 2018;51:80-8. [Crossref] [PubMed]
- Wang Y, Lian B, Si L, et al. Real-world analysis of clinicopathological characteristics, survival rates, and prognostic factors in patients with melanoma brain metastases in China. J Cancer Res Clin Oncol 2021;147:2731-40. [Crossref] [PubMed]
- Govender P, Fashoto SG, Maharaj L, et al. The application of machine learning to predict genetic relatedness using human mtDNA hypervariable region I sequences. PLoS One 2022;17:e0263790. [Crossref] [PubMed]
- Popescu D, El-Khatib M, El-Khatib H, et al. New Trends in Melanoma Detection Using Neural Networks: A Systematic Review. Sensors (Basel) 2022;22:496. [Crossref] [PubMed]
- Agha A, Tarhini AA. Adjuvant Therapy for Melanoma. Curr Oncol Rep 2017;19:36. [Crossref] [PubMed]
- Huang B, Han W, Sheng ZF, et al. Identification of immune-related biomarkers associated with tumorigenesis and prognosis in cutaneous melanoma patients. Cancer Cell Int 2020;20:195. [Crossref] [PubMed]
- Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846-7. [Crossref] [PubMed]
- Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 2017;33:2938-40. [Crossref] [PubMed]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 2019;47:D330-8. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [Crossref] [PubMed]
- Duffy DL, Zhu G, Li X, et al. Novel pleiotropic risk loci for melanoma and nevus density implicate multiple biological pathways. Nat Commun 2018;9:4774. [Crossref] [PubMed]
- De Summa S, Lasorella A, Strippoli S, et al. The Genetic Germline Background of Single and Multiple Primary Melanomas. Front Mol Biosci 2020;7:555630. [Crossref] [PubMed]
- Sumantran VN, Mishra P, Bera R, et al. Microarray Analysis of Differentially-Expressed Genes Encoding CYP450 and Phase II Drug Metabolizing Enzymes in Psoriasis and Melanoma. Pharmaceutics 2016;8:4. [Crossref] [PubMed]
- Rashid G, Bernheim J, Green J, et al. Parathyroid hormone stimulates the endothelial nitric oxide synthase through protein kinase A and C pathways. Nephrol Dial Transplant 2007;22:2831-7. [Crossref] [PubMed]
- Zhang HL, Hu BX, Li ZL, et al. PKCβII phosphorylates ACSL4 to amplify lipid peroxidation to induce ferroptosis. Nat Cell Biol 2022;24:88-98. [Crossref] [PubMed]
- Zhu Z, Yang L, Zhang Y, et al. Increased expression of PRKCB mRNA in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. Ann Hum Genet 2018;82:200-5. [Crossref] [PubMed]
- Kandasamy K, Mohan SS, Raju R, et al. NetPath: a public resource of curated signal transduction pathways. Genome Biol 2010;11:R3. [Crossref] [PubMed]
- Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001;414:799-806. [Crossref] [PubMed]
- Ferrara G, Argenziano G. The WHO 2018 Classification of Cutaneous Melanocytic Neoplasms: Suggestions From Routine Practice. Front Oncol 2021;11:675296. [Crossref] [PubMed]
- Park HY, Perez JM, Laursen R, et al. Protein kinase C-beta activates tyrosinase by phosphorylating serine residues in its cytoplasmic domain. J Biol Chem 1999;274:16470-8. [Crossref] [PubMed]
- Banerjee A, Ray S. Structural Exploration and Conformational Transitions in MDM2 upon DHFR Interaction from Homo sapiens: A Computational Outlook for Malignancy via Epigenetic Disruption. Scientifica (Cairo) 2016;2016:9420692. [Crossref] [PubMed]
- Banerjee A, Ray S. Structural insight with mutational impact on tyrosinase and PKC-β interaction from Homo sapiens: Molecular modeling and docking studies for melanogenesis, albinism and increased risk for melanoma. Gene 2016;592:99-109. [Crossref] [PubMed]
- Voris JP, Sitailo LA, Rahn HR, et al. Functional alterations in protein kinase C beta II expression in melanoma. Pigment Cell Melanoma Res 2010;23:216-24. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


