
Transcriptome analysis of tumor-derived mesenchymal progenitor cells shows that CHST15 is a fibrosis regulator of retroperitoneal liposarcoma
Introduction
Retroperitoneal liposarcoma (RPLS) is a heterogeneous malignancy that accounts for more than 40% of all retroperitoneal sarcoma cases (1). RPLS has distinct clinical characteristics, such as a high rate of local recurrence, repeated relapse after surgery, and relatively less commonly distant metastasis (2). Currently, surgery remains the first-line treatment for both primary and recurrent RPLS (3). There is an urgent demand for effective targeted therapy to treat RPLS (4).Therefore, elucidation of the pathogenesis of RPLS and development of novel therapeutic strategies is essential to improve long-term survival and quality of life of patients with RPLS.
The tumor microenvironment (TME) consists of endogenous host stroma that crosstalk with tumor cells, including mesenchymal progenitor cells (MPCs), inflammatory cells, fibroblasts, and endothelial cells. These components modify tumor growth and development (5). MPCs can differentiate into multiple mesodermal tissue types, including osteoblasts, adipocytes, chondrocytes, and fibroblasts (6). MPCs have been hypothesized to be cells of origin for sarcomas and are recognized as key players in different stages of tumorigenesis (7). Moreover, MPCs may potentially be reprogrammed or can differentiate into fibroblasts. The TME is closely related to the tumor development, so to make it clear of MPCs’ functions and targeting MPCs may control the tumor progression and metastasis (5). Currently, functions and heterogeneity of MPCs in liposarcoma pathology remain unclear (8).
Fibrosis is a pathological hallmark of liposarcoma (9). Liposarcoma cells can trans-differentiate into fibroblasts to generate excess fibrous connective tissue through extracellular matrix deposition (10). Although less explored in TME, fibroblasts may support tumor progression and cause chemoresistance (11). Targeting fibroblasts in liposarcoma may affect liposarcoma treatment (9). Carbohydrate sulfotransferase 15 (CHST15), a specific type II transmembrane glycoprotein enzyme, biosynthesizes chondroitin sulfate E (CS-E) and binds various pathogenic mediators. CHST15/CS-E is capable of fibrogenesis through interacting with fibroblasts and forming collagen fibrils (12). Importantly, research with animal models showed that CHST15 contributes to tissue remodeling during injury and fibrosis in the lung, esophagus, colonand heart (13-17). Furthermore, CHST15 promotes tumor growth and invasion (12). However, potential of CHST15 in diagnosis and pathogenesis of liposarcoma has not been explored. Using transcriptome analysis, we examined the expression of CHST15 in RPLS-derived MPCs and explored its functions in TME. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-963/rc).
Methods
Selection and description of patients
Tissue samples derived from patients with RPLS were obtained at the Department of Retroperitoneal Tumor Surgery, Peking University International Hospital. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Peking University International Hospital (approval No. IRB-2021-079) and informed consent was taken from all the patients. During surgery, liposarcoma tissue and adjacent normal adipose tissue were collected from each patient with an average weight of about 2 g (Table 1). All samples were confirmed by pathology and stored at physiological saline.
Table 1
| Patient no. | Sample no. | Sample type | Gender | Pathological type | Stage | Location |
|---|---|---|---|---|---|---|
| 1 | N3/S3 | Tumor/adjacent adipose tissue | Male | WDLS combined with DDLS | Stage III/G3 | Upper abdomen |
| 2 | S4 | Tumor/adjacent adipose tissue | Female | DDLS | Stage III/G3 | Lower abdomen and pelvic cavity |
| 3 | N8/S8 | Tumor/adjacent adipose tissue | Male | DDLS | Stage III/G3 | Lower abdomen and pelvic cavity |
| 4 | N9/S9 | Tumor/adjacent adipose tissue | Female | DDLS | Stage III/G3 | Left abdomen |
| 5 | N12/S12 | Tumor/adjacent adipose tissue | Male | DDLS | Stage III/G3 | Left abdomen |
WDLS, well-differentiated liposarcoma; DDLS, dedifferentiated liposarcoma.
Isolation and culture of MSCs and MPCs
Normal adipose (n=5) and liposarcoma tissues samples (n=4) were surgically collected from patients with RPLS. Mesenchymal stem cells (MSCs) were isolated from normal adipose tissue, whereas MPCs were isolated from liposarcoma tissue. These samples were segmented and digested with a calibrated digestive solution (0.2% collagenase I, Gibco, CA, USA; MSC basic medium, Clin-Biotech, Beijing, China; 0.25% trypsin, Biological Industries, Kibbutz Beit-Haemek Israel; 1:0.4:0.6) overnight at 37 ℃. Thereafter, samples were treated with 0.05% trypsin (Biological Industries) for 45 min, washed with normal saline solution and centrifuged at 1,000 g for 20 min. The cells were suspended in MSC basic medium, seeded in a six-well plate or T-25 flask at a concentration of 2×105/cm2, and incubated at 37 ℃ containing 5% CO2 until they reached 80% confluence. Then, cells were seeded in T-75 flasks for expansion and harvested for subsequent experiments.
Measurement of cell surface markers
MSCs and MPCs were seeded in six-well plates (1×105 cells/well) and cultured for 48 h. Cells were collected, washed twice with phosphate-buffered saline (PBS), and incubated for 30 min at 4 ℃ in the dark with antibodies according to the manufacturer’s instructions. The fluorescent labeled antibodies were CD90 FITC, CD105 APC, CD34 PE-cy7, CD11b BV605, CD45 FITC, and CD73 PE (BioLegend, San Diego, CA, USA). Then, cells were washed with PBS, centrifuged at 300 g at 4 ℃, resuspended in PBS, and detected and analyzed by flow cytometry (FACSCelestaTM, BD Biosciences, San Diego, CA, USA).
Cell proliferation assay
The Cell Proliferation Dye eFluorTM 670 (Invitrogen, CA, USA) staining assay was conducted to determine proliferation of MSCs and MPCs. MSCs and MPCs were harvested, washed with PBS, and resuspended in PBS. Thereafter, cells were mixed with Dye670 solution to reach a concentration of 10 µM and incubated for 10 min at 37 ℃ in the dark. Labeling was suspended by adding five-fold volumes of complete medium and incubating on ice for 5 min. Immediately, the cells were washed with complete medium thrice and cultured in MSC complete medium at 37 ℃ with 5% CO2 for 0, 24, 48, and 72 h, respectively. At each time point, the cells were collected and fixed with 2% paraformaldehyde in PBS. Cell fluorescence was determined and analyzed using flow cytometry (BD FACSCaliburTM). The proliferation indices were analyzed using Modfit LTTM 5 (Verity Software House, Topsham, ME, USA).
Cell differentiation assays
Upon reaching 100% confluence, the MSC complete medium was replaced with MSC adipogenic or osteogenic differentiation medium (ScienCell, Carlsbad, CA, USA) for 18–21 or 14–16 days, respectively, according to the manufacturer’s instructions. After differentiation, the cells were stained with Oil Red O (Solarbio, Beijing, China) for adipogenic differentiation and Alizarin Red S (Solarbio) for osteogenic differentiation, respectively, according to the manufacturer’s instructions. Thereafter, the cells were photographed using an optical microscope (Leica, Germany).
Transcriptome sequencing of MSCs and MPCs
We selected three groups of paired MSCs and MPCs from 3 patients’ samples. A total of 1×106 cells from each MSC and MPC sample were collected, trypsinized, washed twice with PBS, and frozen in liquid nitrogen. Transcriptome analysis was conducted by Applied Protein Technology Co., Ltd. (Shanghai, China). First, RNA extraction and RNA quality determination was performed. Then, mRNA was enriched by magnetic beads with Oligo (dT), and randomly interrupted by a fragmentation buffer. Using this as a template, the first strand of complementary DNA (cDNA) was synthesized with random hexamers, followed by the addition of dNTPs buffer solution, DNA polymerase I to synthesize the second strand of cDNA, and AMPure XP beads to purify double-stranded DNA. Finally, to repair end fragments and select fragment size, polymerase chain reaction (PCR) enrichment was performed to obtain cDNA library. The cDNA fragments were sequenced using an Illumina® HiSeq instrument.
Untargeted metabolomics of MSCs and MPCs
These 3 MSC and MPC pairs were selected for untargeted metabolomics. A total of 3×106 cells were collected from MSC and MPC samples. The cells were trypsinized, washed twice with PBS, and frozen in liquid nitrogen. Sample extraction was performed according to the manufacturer’s protocol. The samples were separated by Agilent 1290 Infinity LC ultra-high performance liquid chromatography (UHPLC; Agilent, Santa Clara, CA, USA). A TripleTOF® 6600 mass spectrometer (AB SCIEX, Framingham, MA, USA) was used to collect the first and second spectrograms. Finally, XCMS software was used to determine metabolite structure and to analyze differences between metabolites (Applied Protein Technology Co., Ltd.).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted using a MolPure® Cell RNA Kit (Yeasen, Shanghai, China) according to the manufacturer’s instructions. RNA concentration was measured using a spectrophotometer (ACTGENE, NAS-99, Beijing, China). Reverse transcription of 1 µg RNA to cDNA was synthesized using the TransScript® cDNA synthesis kit (Transgen, Beijing, China) according to the manufacturer’s guidelines. The qRT-PCR reaction was carried out using the PerfectStart® Green qPCR SuperMix kit (Transgen) and determined cycle threshold (Ct) values using the Applied Biosystems QuantStudio 3 RT-PCR system (Thermo Fisher Scientific, USA) according to the manufacturer’s guidelines. The total volume of qPCR reaction was 20 µL, which consisted of 1 µL cDNA template, 10 µL supermix, 7.8 µL Rnase-free H2O, 0.4 µL forward primer (10 µM), 0.4 µL reverse primer (10 µM) and 0.4 µL passive reference dye (Transgen). All primers were designed and synthesized by Sangon Biotech (Beijing, China). The primers sequences are listed in Table 2. β-actin was used as an internal control. Differential gene expression was evaluated byCt values and calculated with the 2−ΔΔCt method.
Table 2
| Target | Forward (5'-3') | Reverse (5'-3') |
|---|---|---|
| β-actin | CATGTACGTTGCTATCCAGGC | CTCCTTAATGTCACGCACGAT |
| IL-7R | TGTCGTCTATCGGGAAGGAG | CGGTAAGCTACATCGTGCATTA |
| CHST15 | TCGTGTGGACAGTAAGCAGAT | TGTAAGAAGCCATTACCAAGGTC |
| PKNOX2 | GACGCTGCTGTTTGAGAAATG | ATCGCTGAAGAAGGGTTTGTG |
| ALPL | AACATCAGGGACATTGACGTG | GTATCTCGGTTTGAAGCTCTTCC |
| TGF-β1 | CTAATGGTGGAAACCCACAACG | TATCGCCAGGAATTGTTGCTG |
| TGF-β3 | GGAAAACACCGAGTCGGAATAC | GCGGAAAACCTTGGAGGTAAT |
| α-SMA | GGCATTCACGAGACCACCTAC | CGACATGACGTTGTTGGCATAC |
| FGF2 | AGTGTGTGCTAACCGTTACCT | ACTGCCCAGTTCGTTTCAGTG |
qRT-PCR, quantitative real-time polymerase chain reaction.
Cell cycle assay
The cell cycle assay was conducted using the Cell Cycle Staining Kit (MultiSciences Biotech, Hangzhou, China) according to the manufacturer’s instructions. MSCs and MPCs were collected using trypsin and washed once with PBS. After centrifugation at 300 g, the supernatant was discarded. Then, 1 mL DNA staining solution and 10 µL permeabilization solution was added to the cells and incubated for 30 min in the dark. Then, cells were collected and analyzed using flow cytometry (BD Biosciences). Finally, the cell cycle (G1, S, or G2 phase) was analyzed using Modfit LTTM 5 (Verity Software House).
Immunofluorescence assay
MSCs and MPCs were seeded into confocal Petri dishes. After reaching 80% confluence, cells were washed with PBS and fixed with 4% paraformaldehyde for 15 min. Thereafter, cells were treated with 0.5% Triton X-100 for 20 min. Then, blocking buffer (5% horse serum in PBS) was added and cells were incubated for 30 min. The CHST15 primary antibody (1:100; Proteintech, Rosemont, IL, USA) was added to cells and incubated overnight at 4 ℃. Finally, the Alexa Fluor® 488 fluorescent secondary antibody [goat anti-rabbit IgG (H + L)] at a 1:300 ratio (Invitrogen) was incubated for 1 h and stained with 4',6-diamidino-2-phenylindole (DAPI) for 5 min. Cells were observed under a fluorescence microscope (Leica, Model DMIL).
Plasmid transfection
The CHST15 overexpression plasmid (Transcript sequence: NM_015892) (Genechem Co., Ltd., Shanghai, China) was transfected into MSCs and MPCs using Lipofectamine Stem Transfection Reagent (Invitrogen) according to the transfection protocol. A total of 1.5×105 cells/well were seeded in a six-well plate. When cells reached 30–60% confluence, cells were thoroughly mixed with the Lipofectamine Stem Reagent in Opti-MEMTM medium (Gibco) and plasmid DNA in Opti-MEM medium (Gibco) for 10 min at room temperature. Then, the mixture was added to cells and cultured at 37 ℃ for 1–2 days. Finally, the transfected cells were collected for subsequent experiments.
Enzyme-linked immunosorbent assay (ELISA)
After transfection with the CHST15 overexpression plasmid, the cell culture supernatant was collected to examine the expression of FGF2 using the FGF2 ELISA kit (Jianglai, Shanghai, China). The standards and supernatant were added to the plate in a volume of 50 µL. Then 100 µL HRP conjugated antibody was added to the plate and incubated at 37 ℃ for 1 h. After washing for 5 times, the substrate A and B were added to the plate for 15 min. Finally, stop buffer was added and the signal was detected under 450 nm by a microplate reader (Thermo Fisher Scientific).
Statistical analysis
Data were analyzed using GraphPad Prism version 9.0 software (GraphPad Software, Inc., San Diego, CA, USA). All results are expressed as mean ± standard deviation (SD). Differences between two groups were compared using paired sample t-tests. Statistical significance was set at a P<0.05.
Results
Morphology and osteogenic/adipogenic differentiation of MSCs and MPCs
Both adipose-derived MSCs and liposarcoma-derived MPCs were cultured and identified based on morphology (Figure 1A). More MPCs were positively stained than MSCs (Figure 1B), indicating that MPCs from liposarcoma tissue had stronger potential for osteogenic and adipogenic differentiation than MSCs derived from normal adipose tissue.
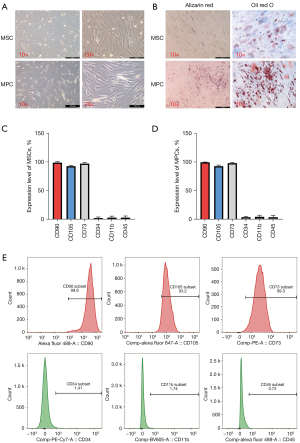
Surface markers of adipose-derived MSCs and liposarcoma-derived MPCs
The MSCs and MPCs were labelled with the standard MSC positive or negative markers, including CD90, CD105, CD73, CD34, CD11b, and CD45 (Figure 1C,1D). All MSCs and MPCs were cultured from different patients’ samples. After incubation, fluorescence was detected by fluorescence-activated cell sorting (FACS). Representative flow charts are shown in Figure 1E. CD90, CD105, and CD73 were positive markers, while CD34, CD11b, and CD45 were negative markers, which was concordant with the standard MSC markers.
MPCs exhibited lower proliferation compared to MSCs
The flow cytometry charts and proliferation indices were analyzed. MSCs and MPCs were arranged in different colors. RPLS-derived MPCs exhibited reduced proliferation than that of adipose tissue MSCs at 24, 48, and 72 h, respectively (P<0.001; Figure 2A,2B).
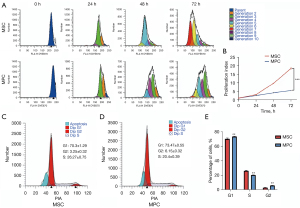
MPCs exhibited longer G1 and G2 phase than MSCs
The cell cycle phases were analyzed using propidium iodide (PI) fluorescence. Flow cytometry indicated that G1 and G2 phases in MPCs were longer than that of MSCs (Figure 2C,2D). The histogram illustrates statistical difference of each phase (P<0.01; Figure 2E). These results correlated with the lower proliferation of MPCs than MSCs.
Transcriptome sequencing of MSCs and MPCs
To compare differential gene expression between MSCs and MPCs, transcriptome sequencing technology was applied to map transcriptional changes in 3 pairs of MSCs and MPCs from 3 patients with RPLS. Then, Padj<0.05, log2fold change (FC) >1 were used as differential significance criterion. A total of 27 differential genes between MPCs and MSCs were identified, including 15 upregulated genes and 12 downregulated genes (Figure 3A). As shown in Figure 3B, IL-7R, PKNOX2, ALPL, CHST15, and MARCHF4 were significantly upregulated, whereas RNF175, DKK2, ITM2A, COL6A6, and DPT were significantly downregulated in MPCs. The Gene Ontology (GO) analysis revealed that MPCs enriched in cell adhesion, biological adhesion, and anatomical structure morphogenesis compared to MSCs (Figure 3C). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database showed that focal adhesion, actin cytoskeleton regulation, and Rap1 signaling pathways were enriched in MPCs (Figure 3D). Additionally, the qPCR results confirmed that IL-7R, PKNOX2, ALPL, and CHST15 were upregulated in MPCs but were not upregulated in MSCs.
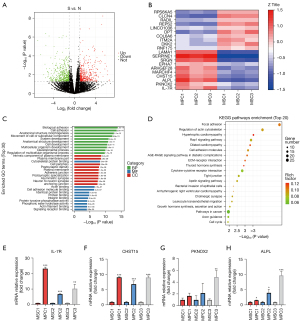
qRT-PCR analysis of upregulated genes by transcriptome sequencing
To verify transcriptome sequencing of upregulated genes, the top expressed genes, including IL-7R, PKNOX2, ALPL, and CHST15 were detected by qRT-PCR. At the mRNA level, these genes were upregulated in MPCs (n=3) but were not upregulated in MSCs (Figure 3E-3H).
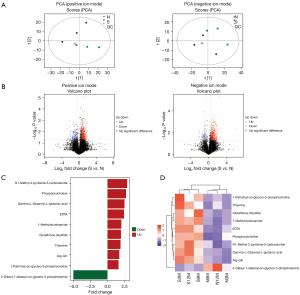
Untargeted metabolomics of MSCs and MPCs
Metabolic differences were determined by untargeted metabolomics using XCMS software. A total of 368 types of metabolites (FC >1.5 or FC <0.67; P<0.05) were found to differ between MSCs and MPCs, of which 272 were in positive-ion mode and 96 were in negative-ion mode. The principal component analysis (PCA) demonstrated a clear difference in quality control (QC) between MSCs and MPCs in positive-ion mode (Figure 4A) and negative-ion mode (Figure 4B). The volcano plot identified significant differences in metabolites in positive and negative ion modes (FC >1.5 or FC <0.67; P<0.05). The significant differences in metabolites analyzed by positive- and negative-ion mode are shown in Figure 4C as a histogram [orthogonal partial least-squares discriminant analysis (OPLS-DA) variable importance projection (VIP) >1; P<0.05]. The metabolites were arranged by FC. N1-methyl-2-pyridone-5-carboxamide, phosphorylcholine, and gamma-L-glutamyl-l-glutamic acid were significantly upregulated in MPCs. The heatmap cluster indicated differential expression levels in different MSC and MPC samples (Figure 4D).
Immunofluorescence staining confirmed CHST15 overexpression in MPCs
The expression of CHST15 in MPCs and MSCs was determined by immunofluorescence staining using Alexa Fluor® 488 fluorescent secondary antibody [goat anti-rabbit IgG (H + L)]. The fluorescence intensity of MPCs was brighter than that of normal MSCs (Figure 5A). This validated the finding that CHST15 had high expression in MPCs.
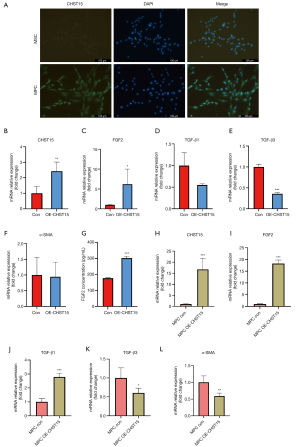
CHST15 gene transfection upregulates FGF2 expression in MSCs and MPCs
To detect fibrosis-related genes (TGF-β1, TGF-β3, α-SMA, and FGF2) at the mRNA level, CHST15 was successfully overexpressed (P<0.01) by plasmid transfection (Figure 5B). Moreover, TGF-β1 and TGF-β3 (P<0.001) was downregulated whereas FGF2 upregulated (P<0.05) compared to the control. However, α-SMA expression was unchanged (Figure 5C-5F). ELISA showed that FGF2 was highly expressed in the supernatant of cell cultures in which CHST15 was overexpressed (P<0.001; Figure 5G). To further confirm the effects of CHST15 on MPCs, CHST15 was successfully overexpressed (P<0.001) in MPCs by plasmid transfection (Figure 5H). FGF2 and TGF-β1 were upregulated (P<0.001), whereas TGF-β3 (P<0.05) and α-SMA (P<0.01) were downregulated (Figure 5I-5L).
Discussion
MPCs are considered as major components and important modulators of TME (18). Novel techniques, including single-cell transcriptome and metabolome, have shown that MPCs display heterogeneity in multiple sources and reprogrammed metabolic characteristics (19,20). The functions of tumor stromal MPCs include exosome secretion, cell fusion and mitochondrial transport to regulate tumor cell growth and proliferation (20-22).
MPCs associated with tumors exhibit different biological properties than normal MSCs and, more importantly, promote tumorigenesis. Demonstrating which cellular signaling pathways are involved in regulating biological processes of MPCs might provide novel approaches for targeted therapy for RPLS. In this study, MPCs were isolated from RPLS patients and adherently cultured. Both normal-adipocyte and tumor-derived MPCs have similar morphologies and expression profiling of surface markers. Moreover, both MSCs and MPCs express CD105, CD73, and CD90. They have the potential for osteogenic and adipocyte differentiation. However, tumor-derived MPCs exhibit a slower growth rate than adipocyte-derived MPCs. This results in an increase in the percentage of cells in the G1 and G2 phases. Therefore, it would be of great interest to characterize biological functions of adipocytes MSCs and tumor-derived MPCs.
Gene sequencing was performed to explore molecular mechanisms underlying how biological functions of MPCs were regulated in RPLS. Transcriptomes exhibit a significant increase in MPCs compared with adipocyte MSCs and are enriched with 15 upregulated genes and 12 downregulated genes. The highly expressed genes in RPLS-derived MPCs included CHST15, IL-7R, PKNOX2, and ALPL. Based on GO and KEGG pathway analyses, these genes were involved in cell adhesion, extracellular matrix, and cell structure formation. Therefore, MPCs may play an important role in TME and may act as a communication link between tumor cells.
RNA sequencing revealed higher expression of CHST15 in liposarcoma stroma-derived MPCs than adipocytes MSCs. CHST15 is a type II transmembrane glycoprotein that regulates CS-E biosynthesis (21). CHST15 inhibition reduced the proliferation and growth of cancer cells both in vitro and in a xenograft model (22). Furthermore, CHST15 overexpression in tumor tissues was associated with poor overall survival and modified identity of epithelial and mesenchymal cells (12,23,24). CHST15-knockdown results in reduced proliferation and enhanced apoptosis by altering CHST15/ILKAP/CCND1 and CHST15/RABL6/PMAIP1 signaling axes in esophageal cancer (25). Furthermore, CHST15 knockdown represses colonic fibrosis and reverses epithelial-mesenchymal transition (26). However, functions of CHST15 in adipocyte MPCs and its association with poor prognosis in patients with liposarcoma need to be elucidated.
In this study, RNA sequencing identified the critical involvement of CHST15 in RPLS. Based on GO analysis, CHST15 was found to be related to the intrinsic component of the plasma membrane. The metabolic differences between MSCs and MPCs were analyzed using untargeted metabolomics. The most related metabolite was gamma-L-glutamyl-L-glutamic acid. Gamma-glutamyl glutamic acid was essential in cystic fibrosis disease (27). Oxidation products of glutathione [glutathione sulfonamide (GSA)] are considered as biomarkers of early cystic fibrosis lung disease (28). Although there is no evidence to confirm that CHST15 overexpression correlates with aberrant metabolic levels, the overexpressed CHST15 is proposed to participate in fibrosis of RPLS.
Currently, using immunostaining for CHST15 in clinical evaluation and diagnosis remains challenging. However, our results indicate that a high expression of CHST15 correlates with fibrosis and prognosis of liposarcoma. Existing research has reported that CHST15 is associated with breast cancer (24), pulmonary metastasis, and tissue fibrosis (13). Using CHST15-overexpression plasmid-mediated gene transduction, CHST15 expression is required for the activation of fibrosis. Furthermore, CHST15 transduction leads to upregulation of FGF2, which may promote fibrosis in TME. Therefore, CHST15 regulates the activation of fibrosis in RPLS multiple progenitor cells, which may provide a novel and targeted therapeutic strategy to treat RPLS.
Acknowledgments
Funding: This work was supported by the National Key Research and Development Program of China (No. 2017YFC1200503), the Key Research and Development Program of Hebei Province (No. 19272405D), the Natural Science Foundation of Shandong Province (No. ZR2020MH327), the Peking University International Hospital Research Fund (No. YN2021ZD04), and CAS “Light of West China Program” in 2018.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-963/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-963/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-963/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethical Committee of Peking University International Hospital (approval No. IRB-2021-079) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Improta L, Tzanis D, Bouhadiba T, et al. Overview of primary adult retroperitoneal tumours. Eur J Surg Oncol 2020;46:1573-9. [Crossref] [PubMed]
- Cheung KT, Mitchell C, Wong E. Retroperitoneal liposarcoma in a nonagenarian. Autops Case Rep 2021;11:e2020224. [Crossref] [PubMed]
- Carbone F, Pizzolorusso A, Di Lorenzo G, et al. Multidisciplinary Management of Retroperitoneal Sarcoma: Diagnosis, Prognostic Factors and Treatment. Cancers (Basel) 2021;13:4016. [Crossref] [PubMed]
- Dumitra S, Gronchi A. The Diagnosis and Management of Retroperitoneal Sarcoma. Oncology (Williston Park) 2018;32:464-9. [PubMed]
- Sounni NE, Noel A. Targeting the tumor microenvironment for cancer therapy. Clin Chem 2013;59:85-93. [Crossref] [PubMed]
- Jackson WM, Nesti LJ, Tuan RS. Potential therapeutic applications of mus-cle-derived mesenchymal stem and progenitor cells. Expert Opin Biol Ther 2010;10:505-17. [Crossref] [PubMed]
- Barry FP, Murphy JM. Mesenchymal stem cells: clinical applications and biological characterization. Int J Biochem Cell Biol 2004;36:568-84. [Crossref] [PubMed]
- Takeuchi S, Yamanouchi K, Sugihara H, et al. Differentiation of skeletal muscle Mesenchymal progenitor cells to myofibroblasts is reversible. Anim Sci J 2020;91:e13368. [Crossref] [PubMed]
- Si HP, Wang Z, Fan QH, et al. Dedifferentiated liposarcoma with inflammatory my-ofibroblastic tumor-like features: a clinicopathological analysis of five cases. Zhonghua Bing Li Xue Za Zhi 2019;48:282-7. [PubMed]
- Bovée JVMG. Jason L. Hornick: Practical soft tissue pathology: a diagnostic ap-proach, 2nd edition. Virchows Arch 2018;473:785-6.
- Asanuma K, Matsumine A, Nakamura T, et al. Impact of plasma fibrinogen levels in benign and malignant soft tissue tumors. Cancer Biomark 2016;16:453-8. [Crossref] [PubMed]
- Matsuda Y, Fujii Y, Matsukawa M, et al. Overexpression of carbohydrate sulfotrans-ferase 15 in pancreatic cancer stroma is associated with worse prognosis. Oncol Lett 2019;18:4100-5. [PubMed]
- Kai Y, Tomoda K, Yoneyama H, et al. Silencing of Carbohydrate Sulfotransferase 15 Hinders Murine Pulmonary Fibrosis Development. Mol Ther Nucleic Acids 2017;6:163-72. [Crossref] [PubMed]
- Watanabe K, Arumugam S, Sreedhar R, et al. Small interfering RNA therapy against carbohydrate sulfotransferase 15 inhibits cardiac remodeling in rats with dilated cardiomyopathy. Cell Signal 2015;27:1517-24. [Crossref] [PubMed]
- Suzuki K, Yoneyama H. New endoscopic approach of anti-fibrotic therapy for in-flammatory bowel disease. Ann Transl Med 2017;5:191. [Crossref] [PubMed]
- Sato H, Sagara S, Nakajima N, et al. Prevention of esophageal stricture after endo-scopic submucosal dissection using RNA-based silencing of carbohydrate sul-fotransferase 15 in a porcine model. Endoscopy 2017;49:491-7. [Crossref] [PubMed]
- Suzuki K, Yokoyama J, Kawauchi Y, et al. Phase 1 Clinical Study of siRNA Target-ing Carbohydrate Sulphotransferase 15 in Crohn's Disease Patients with Active Mu-cosal Lesions. J Crohns Colitis 2017;11:221-8. [Crossref] [PubMed]
- Wu C, Amini-Nik S, Nadesan P, et al. Aggressive fibromatosis (desmoid tumor) is derived from mesenchymal progenitor cells. Cancer Res 2010;70:7690-8. [Crossref] [PubMed]
- Kieffer Y, Hocine HR, Gentric G, et al. Single-Cell Analysis Reveals Fibroblast Clusters Linked to Immunotherapy Resistance in Cancer. Cancer Discov 2020;10:1330-51. [Crossref] [PubMed]
- Davidson S, Efremova M, Riedel A, et al. Single-Cell RNA Sequencing Reveals a Dynamic Stromal Niche That Supports Tumor Growth. Cell Rep 2020;31:107628. [Crossref] [PubMed]
- Cheung ST, Miller MS, Pacoma R, et al. Discovery of a Small-Molecule Modulator of Glycosaminoglycan Sulfation. ACS Chem Biol 2017;12:3126-33. [Crossref] [PubMed]
- Takakura K, Shibazaki Y, Yoneyama H, et al. Inhibition of Cell Proliferation and Growth of Pancreatic Cancer by Silencing of Carbohydrate Sulfotransferase 15 In Vitro and in a Xenograft Model. PLoS One 2015;10:e0142981. [Crossref] [PubMed]
- Bhattacharyya S, Feferman L, Han X, et al. Increased CHST15 follows decline in arylsulfatase B (ARSB) and disinhibition of non-canonical WNT signaling: poten-tial impact on epithelial and mesenchymal identity. Oncotarget 2020;11:2327-44. [Crossref] [PubMed]
- Liu LC, Wang YL, Lin PL, et al. Long noncoding RNA HOTAIR promotes invasion of breast cancer cells through chondroitin sulfotransferase CHST15. Int J Cancer 2019;145:2478-87. [Crossref] [PubMed]
- Wang X, Cheng G, Zhang T, et al. CHST15 promotes the proliferation of TE-1 cells via multiple pathways in esophageal cancer. Oncol Rep 2020;43:75-86. [PubMed]
- Suzuki K, Arumugam S, Yokoyama J, et al. Pivotal Role of Carbohydrate Sul-fotransferase 15 in Fibrosis and Mucosal Healing in Mouse Colitis. PLoS One 2016;11:e0158967. [Crossref] [PubMed]
- Masood A, Jacob M, Gu X, et al. Distinctive metabolic profiles between Cystic Fi-brosis mutational subclasses and lung function. Metabolomics 2021;17:4. [Crossref] [PubMed]
- Dickerhof N, Turner R, Khalilova I, et al. Oxidized glutathione and uric acid as bi-omarkers of early cystic fibrosis lung disease. J Cyst Fibros 2017;16:214-21. [Crossref] [PubMed]
(English Language Editor: C. Mullens)


