
Involvement of NEAT1/PINK1-mediated mitophagy in chronic obstructive pulmonary disease induced by cigarette smoke or PM2.5
Introduction
Chronic obstructive pulmonary disease (COPD) is a common clinical chronic respiratory disease, with incompletely reversible and persistent airflow limitations (1-3). In China, the prevalence of COPD in the elderly is about 13.6% (2), and increases with age (1,2). Cigarette smoking (CS), and ambient air pollution, mainly including fine particular matter (PM2.5), are the main risk factors for COPD (4,5). Recent studies have indicated that these risk factors affect mitochondrial function and morphology (6-9) in the occurrence and development of COPD. Mitophagy is a highly conserved and selective form of autophagy that eliminates dysfunctional mitochondria, and is mainly governed by the PINK1/Parkin pathway. Mitophagy has possessed a dual role in COPD (7,10-12), contributing to the pathogenesis of COPD. In bronchial epithelial cells of COPD patients, an increased PTEN-induced kinase 1 (PINK1) and decreased Parkin indicated an insufficient mitophagy through accumulation of damaged mitochondria (11). On the other side, it was found that PINK1−/− mice were resistant to emphysema and protect ed against mitochondrial dysfunction after CS exposure (10), suggesting PINK1-dependent mitophagy have been implicated in COPD development (7,11), however, it is still unknown which the level of PINK1 has been regulated.
Long non-coding ribonucleic acids (lncRNAs) are greater than 200 nucleotides, have no protein coding functions, and are widely involved in biological processes (13,14). Most lncRNAs have been implicated in the occurrence and development of COPD (15,16). LncRNA nuclear enriched abundant transcript 1 (NEAT1) is thought to be involved in a variety of cancers and inflammatory diseases (17-19). NEAT1 was found to be upregulated in COPD patients, and NEAT1 expression could predict decreased COPD susceptibility and acute exacerbation risk, and negatively correlated with GOLD stage and levels of TNF-α, IL-1β, IL-6 and IL-17 in both Acute Exacerbations of COPD (AECOPD) and stable COPD patients (20,21); however, the underlying mechanisms are still not clear. NEAT1 might be involved in the quality control of mitochondria and have the capability to cross-regulate paraspeckles and mitochondria (22), and NEAT1 might interact with PINK1 by stabilizing PINK1 ubiquitination and degradation (23,24) and promoting mitophagy (23) in the development of Parkinson’s disease. However, there is a lack of evidence about the relationship between upregulated NEAT1 and PINK1-mediated mitophagy in development of COPD. Generally, dysregulated lncRNAs respond to environmental stimuli; for example, the overexpression of maternally expressed 3 (MEG3) was found in Primary Human Pulmonary Microvascular Endothelial Cells (HPMEC) induced by CS extract (CSE), and cell apoptosis was increased (25). lung cancer progression-associated transcript 1 (LCPAT1) was increased in lung cancer cell lines exposed to CSE and PM2.5 (26). However, the change of NEAT1 expression induced by CS and PM2.5 remains poor.
This study, we for the first time sought to clarify the relationship between NEAT1 and PINK1/Parkin-mediated mitophagy in COPD pathogenesis, and explore their roles in COPD induced by CS and PM2.5. Thus, this study attempted to investigate the effect of NEAT1 in a COPD animal model and determine whether its expression was associated with PINK1/Parkin-mediated mitophagy. Further, the involvement of NEAT1 and PINK1/Parkin-mediated mitophagy in COPD pathogenesis was examined using the small interfering RNA (siRNA) NEAT1 in vitro. Thus, this work revealed that the NEAT1/PINK1 pathway upregulated in COPD rat model and in A549 cell exposed by CS and PM2.5, suggesting a new light for pathogenesis of COPD. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-542/rc).
Methods
Preparation of PM2.5 and CSE
PM2.5 samples were obtained using an intelligent PM2.5-TSP medium flow sampler (Zhengzhouhuazhi, Zhengzhou, China) in Putian, China, and collected using an ultrafine glass fiber filter. The PM2.5 samples were eluted with sterilized water from filters, and freeze-dried to produce PM2.5 powders. The powders were then observed by scan electron microscopy (SEM; SU8010, Hitachi, Japan), and the hydrodynamic size and Zeta potential of PM2.5 were detected by a Zetasizer (Malvern Nano-ZS90, UK). The PM2.5 samples were stored at −80 ℃. As in a previous study (27), 100% CSE was extracted from 6 cigarettes (Fujian China Tobacco Industry Co., Ltd; flue-cured tobacco type, smoke nicotine content: 0.7 mg, tar content: 10 mg, smoke carbon monoxide content: 12 mg), which was bubbled through 30 mL of Roswell Park Memorial Institute (RPMI)-1640 (Hyclone, American). The solution was adjusted to pH 7.4 with 1 M of sodium hydroxide (NaOH), and then sterilized using a 0.22-µm microporous membrane filter, and stored at −80 ℃.
Animal and treatment
Wistar rats were purchased from Shanghai SLAC Laboratory Animal Co., China [Batch: SCXK(Hu) 2017-0005]. The 30 male rats, aged 6–8 weeks, weighted 180±20 g, were divided into a CS exposure (CSM) group, a PM2.5 exposure (PMM) group, and a control (Ctrl) group using a random number table. The rats were housed in an animal room at Putian University, placed on a 12:12 h light/dark cycle, and given free access to food and water. As per previous literature (28), the CSM model was established by chronic and passive smoking exposure, the PMM model was established through PM2.5 suspension passive atomization exposure using a 405C Medical Nebulizer (Yuyue, China), and the Ctrl group received a 0.9% sodium chloride injection (Anhui Fengyuan, China). The process of the exposure was continuous for 90 days, and the activity state of the experimental rats was closely observed (see Figure S1). Experiments were performed under a project license (No. 201924) granted by the ethics committee of the Affiliated Hospital of Putian University, in compliance with Chinese guidelines (GB/T 35892-2018) for the care and use of animals.
Pulmonary function detection
The pulmonary function of the COPD model was analyzed by RM6240B multi-channel physiological signal acquisition and a processing system. After exposure, the rats were anesthetized with an intraperitoneal injection of 3% Pentobarbital (0.1 mL/100 g). The neck of the rats was cut open, and the main bronchus was carefully separated. Next, the T-shaped mouth was cut out using ophthalmic scissors carefully in the main bronchus, and a HX200 respiratory flow transducer was connected for endotracheal intubation using channel 1 of a RM6240B multi-channel physiological signal acquisition and processing system. The tidal volume (VT), respiratory rate (f), and minute ventilation (MV) was then measured using the physiological signal processing system.
ELISA analysis of serum and BALF
The blood of each rat was drawn from the abdominal aorta into an Eppendorf (EP) tube pre-treated with Ethylenediaminetetraacetic acid (EDTA). The serum was extracted after centrifugation, and stored at −80 ℃. After the rats were sacrificed, the left lungs were lavaged 3 times with 2 mL of phosphate buffered solution, and the lung lobes were gently massaged, and bronchoalveolar lavage fluid (BALF) was collected. The levels of inflammatory cytokines in the serum and BALF, including the interleukin (IL)-6, IL-8 and tumor necrosis factor alpha (TNF-α) levels, were determined using enzyme-linked immunoassays (ELISAs) (Wuhan Hengyuan, China) in strict accordance with the manufacturer’s instructions.
Histology and IHC evaluations
Fresh animal lung tissue was taken and fixed in 10% neutral formalin for 48 h. Pathological sections of the lung tissues were fixed and stained with hematoxylin and eosin (H&E). The 5.0-µm thick slides were mounted with neutral gum, and evaluated under a microscope (DM1L, Leica, Germany). Next, the lung sections were deparaffinized and incubated with a MUC5AC antibody (1:200, GeneTex, USA), PINK1 antibody (1:200, Proteintech, China), and Parkin antibody (1:200, Immunoway, USA). Subsequently, the sections were incubated with the Horseradish Peroxidase (HRP)-conjugated anti-rabbit immunoglobulin G (IgG), and counterstained with hematoxylin in accordance with the manufacturers’ instructions. The images were captured and evaluated using Fiji software.
TEM observations
The fresh lung samples were quickly cut into pieces of about 1 mm3, and the samples were then fixed with 2.5% glutaraldehyde solution for 24 h. Next, 1% osmic acid solution was added to the samples for 2 h, and they were then dehydrated with ethanol (50–100%). Subsequently, the samples were embedded with epoxy resin Spurr, and cut into ultra-thin sections. The sections were observed to examine the ultrastructures of the lung tissues under a transmission electron microscope (TEM; TECNAI G2, FEI, USA).
Cell cultures and treatments
The A549 human alveolar basal epithelial cell line was purchased from Procell, Wuhan, China. The A549 cells were cultured in RPMI 1640 (Hyclone, USA) containing 10% fetal bovine serum (Gibco, USA) and 1% Penicillin-Streptomycin (Hyclone, USA), and incubated at 37 ℃ in a 5%-carbon dioxide incubator. The culture medium was renewed every day. The PM2.5 was suspended by RPMI-1640 and adjusted to 200 µg/mL (PMS), and 10% CSE was diluted by RPMI-1640 (see Figure S2). The A549 cells, which had been cultured and had grown by 70–80%, were treated with the PMS and 10% CSE for 24 h, meanwhile normal cultured were used as controls (CON). Next, for the internal mechanism exploration, the A540 cells were first cultured until they had grown by about 50%, and then transiently transfected with siRNA NEAT1 (si-NEAT1, F: 5'-GUGAGAAGUUGCUUAGAAATT-3'; R: 5'-UUUCUAAGCAACUUCUCACTT-3') or the control siRNA (si-NC, F: 5'-UUCUCCGAACGUGUCACGUTT-3'; R: 5'-ACGUGACACGUUCGGAGAATT-3') for 24 h. Next, the transfected A549 cells were treated with PMS and 10% CSE and incubated for 24 h.
MMP assays
When mitochondria are damaged, the mitochondrial membrane potential (MMP) usually decreases. In this study, the JC-1 probe (Solarbio, China) was used to assess MMP. The JC-1 working solution was incubated with the treated A549 cells for 20 min, and images were captured using a fluorescence microscope (Nikon, Japan). The red/green fluorescence intensity ratio was then calculated using Fiji software.
Mitophagy observation
Mitophagy was detected by the colocalization of mitochondria with lysosome. In this study, the treated A549 cells were incubated with the MitoTracker Red CMXRos Probe (200 nM; Yeasen, China) for 20 min, and incubated with the LysoTracker Green DND-26 Probe (66.7 nM; Yeasen, China) for 45 min. Images were taken with a laser scanning confocal microscope (Nikon C2, Tokyo, Japan).
qRT-PCR detection
Total RNA was extracted from the fresh lung tissue of rats and the treated A549 cells using TRIzol (Invitrogen, American) in accordance with the manufacturer’s instructions. The primer sequences used for the rats were as follows: NEAT1 (F: 5'-TGGCTAGCTCAGGGCTTCAG-3'; R: 5'-TCTCCTTGCCAAGCTTCCTTC-3'), as per previous studies (29,30), and β-actin (F: 5'-CGAGTACAACCTTCTTGCAGC-3'; R: 5'-ACCCATACCCACCATCACAC-3') served as the housekeeping control. Second, for the total RNA of the treated A549 cells, the primer sequences were as follows: NEAT1 (F: 5'-GACCATAGATGAGCAACGGAG-3'; R: 5'-GCATAGCCACTCATCCCATT-3'), and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (F: 5'-CGAGTACAACCTTCTTGCAGC-3'; R: 5'-ACCCATACCCACCATCACAC-3'). Finally, the expression of NEAT1 was examined using BeyoFast™ SYBR Green qPCR Mix (Beyotime, China) on the ABI 7500 Real-Time PCR System (ThermoFisher, USA), and the relative expression level was calculated using the 2−ΔΔCt method.
Western blotting
The protein extracts of the lung tissues and cells were prepared using Radio Immunoprecipitation Assay (RIPA) buffer (Phygene, China) with Phenylmethanesulfonyl fluoride (PMSF) (Solarbio, China), Next, the protein concentrations were tested by a protein quantification kit (BCA Assay) (Zomanbio, China). The added protein was loaded equally, separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (Beyotime, China), and transferred to polyvinylidene fluoride membranes (Beyotime, China). After blocking with 5% non-fat milk, the membranes were incubated with the following primary antibodies: LC3B (1:1,000, Immunoway, USA), p62/SQSTM1 (1:1,000, Proteintech, China), PINK1 antibody (1:1,000, Proteintech, China), Parkin antibody (1:1,000, Immunoway, USA), and GAPDH (1:1,000, Beyotime, China). They were then incubated with a secondary antibody (HRP-conjugated anti-rabbit IgG). The signals of the protein bands were detected by a UVP imaging system (UVP, USA) and assessed using Fiji software.
Statistical analysis
The experimental data were recorded using WPS Office (3.0), and the statistical analysis and plots were conducted with the R language (4.0) packages of tidyverse, Rmisc, ggplot2, ggsignf, and other packages. All the data are expressed as the mean ± standard error of the mean (SEM). The differences between different groups were analyzed with a 1-way analysis of variance or Kruskal-Wallis test. Pairwise comparisons were performed by the Tukey post-hoc test or Nemenyi test. A P value <0.05 was considered statistically significant.
Results
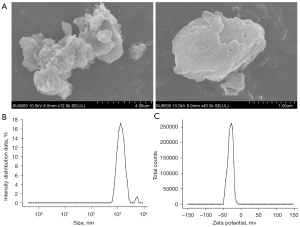
Characterization of PM2.5
The form and size of the PM2.5 that collected were scanned by SEM (see Figure 1A), and were observed to be irregular in shape and variable in size. The hydrodynamic diameter and Zeta potential determination showed that PM2.5 maintained a relatively stable dispersity in normal saline (NS) (see Figure 1B,1C).
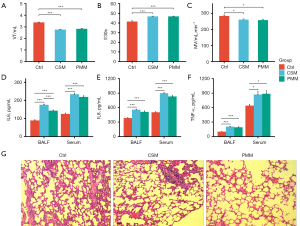
Pulmonary injury of experimental rats
The development of respiratory disease can be evaluated by pulmonary function. In this study, the CSM and PMM groups had significantly decreased VT and MV, and increased f compared to the Ctrl group (P<0.05; see Figure 2A-2C). Inflammatory cytokines are key indicators in COPD, and as Figure 2D-2F show, the IL-6, IL-8, and TNF-a levels in the serum and BALF of the CSM and PMM groups were higher than those of the Ctrl group (P<0.05). In relation to pulmonary function and inflammatory cytokines, no significant difference was found between the CSM and PMM groups (P>0.05).
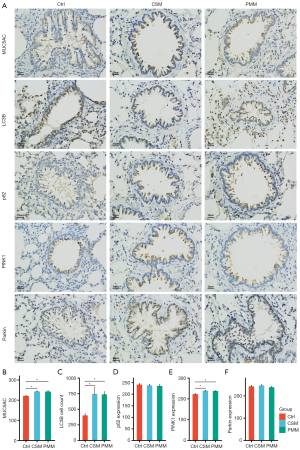
Histological changes in the lung tissue of the experimental rats
As Figure 2G shows, histological alterations in the lung tissues of the different groups were observed. In the Ctrl group, the alveolar size was relatively uniform, the alveolar wall was relatively intact, and no obvious inflammatory cell infiltration was observed. Conversely, both the CSM and PMM groups displayed alveolar enlargement, alveolar wall thinning, alveolar septum rupture, emphysematous bullae and inflammatory responses. Thus, airway remodeling, pulmonary bullae formation, and emphysema appeared to occur in both the CSM and PMM groups. Mucus hypersecretion is a critical histological marker in COPD models (31). Thus, the expression of MUC5AC in lung tissue sections was detected through IHC, and the expression of MUC5AC was increased in both the CSM and PMM groups compared to the Ctrl group (P<0.05; see Figure 3A,3B). These results indicated that COPD models induced by CS and PM2.5 exposure were successfully established.
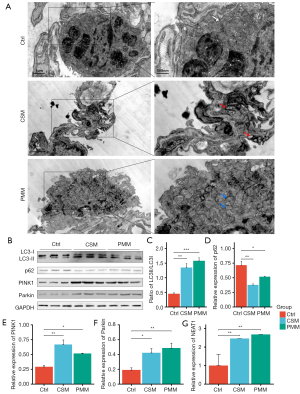
Mitophagy alteration in experimental rats
PINK1/Parkin-mediated mitophagy is an essential mechanism of COPD (7,10,32). In this study, the levels of LC3B and p62 in the lung tissues of the COPD rats were evaluated using IHC images. First, the LC3B cell count was measured, and the counts of both the CSM and PMM groups were higher than that of the Ctrl group (P<0.05; see Figure 3A,3C). Second, there were no significant differences between the 3 groups in terms of p62 levels (P=0.78; see Figure 3A,3D). Third, the expression of PINK1 and Parkin were detected, and both the CSM and PMM groups showed upregulated PINK1 (P<0.05; see Figure 3A,3E). There was no significant difference in the expression of Parkin (P=0.58; see Figure 3A,3F). In relation to the ultrastructure observations, the TEM images of the lung tissues of the 3 groups showed that both the CSM and PMM groups had more alveolar epithelial cell mitochondrial swelling and vacuole formation than the Ctrl group. Additionally, autophagosomes were observed in the CSM and PMM groups (see Figure 4A).
Dysregulation of the NEAT1/PINK1 pathway in the experimental rats
NEAT1 plays an important role in COPD (20); however, the relationship of NEAT1 and PINK1 in the pathogenesis of COPD is unclear. In this study, the levels of LC3B, p62, PINK1, and Parkin were detected by Western blot assays. As Figure 4B shows, compared to the Ctrl group, the LC3 II/I ratios of both the CSM and PMM groups were increased (P<0.05, see Figure 4C), but the expression level of p62 was decreased (P<0.05, see Figure 4D), which suggests enhanced autophagy in the lung tissue of the COPD rats. The expression levels of PINK1 and Parkin in both the CSM and PMM groups were increased (P<0.05, see Figure 4E,4F). Additionally, the expression levels of NEAT1 in both the CSM and PMM groups were significantly increased (see Figure 4G). In conclusion, CS and PM2.5 promoted the expression of NEAT1 and enhanced PINK1/Parkin-mediated mitophagy in COPD rats. However, the regulation of NEAT1 and PINK1/Parkin-mediated mitophagy requires further study.
The regulation of NEAT1 and PINK1 in vitro
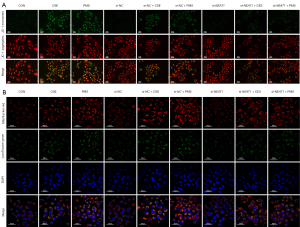
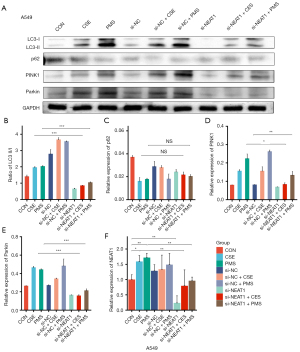
To explore the relationship between NEAT1 and PINK1/Parkin-mediated mitophagy in vitro, the level of NEAT1 was knocked down by si-NEAT1. Notably, after 10% CSE and PMS treatment, the level of MMP decreased. Thus, the knockdown of NEAT1 improved the level of MMP (see Figure 5A). Next, we conducted a confocal microscopy analysis to observe the mitophagy of cells (see Figure 5B), and found that 10% CSE and PMS enhanced mitophagy. Specifically, compared to CON, there were more green fluorescent dots in the A549 cells treated by 10% CSE and PMS. The green fluorescent dots should have been reduced in the A549 cells pretreated with si-NEAT1 and exposed to 10% CSE and PMS. The images indicated that the downregulation of NEAT1 lessened mitophagy. Next, we examined changes in mitophagy-related protein levels (see Figure 6), and found that 10% CSE or 200 µg/mL of PMS increased the ratio of LC3 II/I and decreased the expression level of p62, which suggests that CSE and PMS enhance autophagy. The A549 cells pretreated with si-NEAT1 and exposed to CSE or PMS also had a relatively lower ratio of LC3-II/I and a higher level of p62 than the A549 cells that were not pretreated. Next, the expression level of PINK1 and Parkin in the A549 cells exposed to 10% CSE and PMS was increased, and relatively decreased after the downregulation of NEAT1. Taken together, these results indicate that the NEAT1/PINK1 pathway plays an important role in CS and PM2.5 induced mitochondrial damage and mitophagy disorder, and in the occurrence and development of COPD.
Discussion
The prevention and control of COPD have become a public health priority in China, and are listed as health actions in China [2019–2030]. Due to rapid industrialization and urbanization, ambient air pollution has become a secondary risk factor for COPD in China (1,17,33). PM2.5 is a main component of ambient air pollution, and induces and accelerates COPD (34-36); however, the mechanisms involved in the occurrence and development of COPD induced by environmental risk factors are still being investigated (37-39). Traditionally, studies on the pathogenesis of COPD have often used CS exposure in vivo and in vitro. In recent years, PM2.5 exposure alone or combined with CS exposure has been used to establish COPD models (28), which can be used to explore the role of the environmental risk factors in the occurrence and development of COPD. In this study, COPD rat models induced by both CS and PM2.5 exposure were successfully established, and functional and histological pulmonary injuries in the COPD models were clearly observed. The mitophagy disorder is a biological characterization of, and a regulation mechanism involved in, the pathogenesis of COPD (8,10). In this study, mitochondrial dysfunctions and autophagosomes in the lung tissues of the 2 rat model were observed in TEM images. Additionally, consistent with recent studies (7,10,11), the levels of PINK1 increased, the ratios of LC3-II/I increased, and the levels of p62 decreased in the COPD rat models.
Mitophagy provides a main pathway for eliminating damaged and depolarized mitochondria (32,40,41), and plays an important role in the mitochondrial quality control of COPD (8). PINK1/Parkin pathway is one of mechanisms of mitophagy, PINK1 regulates mitochondrial integrity and functions and combines with Parkin, which is downstream of PINK1 and amplifies the mitophagy signal (42,43). Recent studies (10,44) and this study found that the expression of PINK1 was increased in COPD models induced by CS and PM2.5. Mizumura et al. illustrated that abnormal mitochondria, tracheal lumen dilation, and abnormal lung cilia clearance are reduced in PINK1−/− mice exposed to CS (10). Further, Mannam et al. found that the expression of PINK1 and Parkin synergistically increased in the lung tissues of mouse exposed to CS (45). These data suggest that mitochondrial depolarization, mitochondria damage, and PINK1/Parkin-mediated mitophagy activation are related to the progression of COPD (12).
In addition to enhanced mitophagy, insufficient mitophagy was also observed to be involved in the progression of COPD. Ito et al indicated that the high expression of PINK1 in the lung homogenates of COPD patients, which might be related to aggregate damaged mitochondria (11). Additionally, impaired Parkin translocation to damaged mitochondria has been observed in the lung tissues of COPD patients and smokers (44), and it showed that the level of Parkin might be correlated with pulmonary function of COPD patients (11). In vitro, CSE reduces Parkin translocation to damaged mitochondria regardless of the level of PINK1, which suggests that Parkin serves as a rate limiting factor of PINK1/Parkin-mediated mitophagy (7). Both CS and PM2.5 had an adverse effect on mitochondria. When BEAS-2B cells were exposed to PM2.5, MMP collapsed, the Reactive oxygen species (ROS)/mitochondrial ROS (mtROS) level increased, and cellular respiration was suppressed (6,46). This study also found that CS and PM2.5 damaged mitochondria, reduced MMP, and enhanced PINK1/Parkin-mediated mitophagy, but the underlying mechanisms are not yet fully understood.
Notably, various lncRNAs, such as MALAT1 (47), MIR155HG (48), and NEAT1 (20), are associated with respiratory diseases, and are widely involved in the toxicity of CS and PM2.5. Previous studies have shown that the level of NEAT1 was increased in COPD and AECOPD patients, and was correlated with the stage of COPD (20,21). Research has shown that NEAT1 promotes PINK1 ubiquitination and degradation and impairs PINK1-dependent mitophagy (23,24). However, the role of upregulated NEAT1 in COPD is still unclear. In this study, we found that the expression of NEAT1 and the level of PINK1 were elevated in vivo, which suggests that they may be closely related to the pathological changes of COPD. To determine the regulated mechanism of NEAT1 and PINK1 in COPD, si-NEAT1 was simulated in the A549 cells, and PINK1-mediated mitophagy alterations after exposure to 10% CSE and PMS were observed. When NEAT1 was knocked down, mitophagy activation and MMP reduction induced by CSE and PMS were significantly decreased under fluorescence observation. Subsequently, the mitophagy-related proteins LC3B, p62, PINK1, and Parkin, were detected by Western blotting. The level of PINK1 and the ratio of LC3-II/I both changed with the expression of NEAT1. These results indicate that NEAT1 might promote PINK1-mediated mitophagy and play an important role in COPD pathogenesis.
This study was the first to explore the role of the upregulated expression of NEAT1 in the occurrence and development of COPD. In vivo, we found a functional correlation between the abnormal expression of NEAT1 and PINK1-mediated mitophagy in COPD following CS and PM2.5 exposure. Additionally, in vitro, upregulated NEAT1 expression was positively correlated with the mitochondria damage of the lung epithelial cells and with the increased expression of PINK1. Thus, there is a regulatory mechanism between NEAT1 and PINK1-mediated mitophagy.
This study had some limitations. First, like previous studies (28,49), this study constructed COPD experiment models induced by CS and PM2.5, but the COPD models did not fully be consistent with reality. For example, the exposed concentration of PM2.5 was not easy to assess, and might be higher than that in the actual environment. Second, this study only focused on mitochondrial dysfunctions and the role of mitophagy; however, mitophagy is a complicated process regulated by multiple pathways and factors (12,50,51). This study did not examine other pathways due to experimental condition limitations. Third, NEAT1 was proven to be involved in the regulation of PINK1/Parkin-mediated mitophagy in COPD, but the regulated mechanism requires further research.
In conclusion, this study showed that exposure to CS and PM2.5 can be used to successfully establish COPD rat models, and caused mitochondrial dysfunctions and mitophagy disorder in vivo and in vitro. The upregulation of NEAT1 was involved in CS/PM2.5-induced lung tissue injury and bronchial epithelial cell damage with PINK1/Parkin-mediated mitophagy activation. Most importantly, CS and PM2.5 increased the expression of NEAT1 to upregulate PINK1 level and enhance mitophagy in the progression of COPD. Thus, NEAT1 and PINK1 might be potential therapeutic targets and prognosis biomarkers in COPD.
Acknowledgments
Funding: This study was supported by the General Program of the Natural Science Foundation of Fujian Province, China (No. 2020J011251); the Fujian Health Provincial Technology Project, China (No. 2020GGA078); the Open Project of the Key Laboratory of Environment and Health of Fujian Province, China (No. GWGXZD-201801); and the Science and Technology Project of Putian City, China (No. 2019S3F002).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-542/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-542/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-542/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (No. 201924) granted by the ethics committee of the Affiliated Hospital of Putian University, in compliance with Chinese guidelines (GB/T 35892-2018) for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang C, Xu J, Yang L, et al. Prevalence and risk factors of chronic obstructive pulmonary disease in China (the China Pulmonary Health [CPH] study): a national cross-sectional study. Lancet 2018;391:1706-17. [Crossref] [PubMed]
- Fang L, Gao P, Bao H, et al. Chronic obstructive pulmonary disease in China: a nationwide prevalence study. Lancet Respir Med 2018;6:421-30. [Crossref] [PubMed]
- GBD Chronic Respiratory Disease Collaborators. Prevalence and attributable health burden of chronic respiratory diseases, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir Med 2020;8:585-96. [Crossref] [PubMed]
- Manisalidis I, Stavropoulou E, Stavropoulos A, et al. Environmental and Health Impacts of Air Pollution: A Review. Front Public Health 2020;8:14. [Crossref] [PubMed]
- Zhao J, Li M, Wang Z, et al. Role of PM2.5 in the development and progression of COPD and its mechanisms. Respir Res 2019;20:120. [Crossref] [PubMed]
- Qiu YN, Wang GH, Zhou F, et al. PM2.5 induces liver fibrosis via triggering ROS-mediated mitophagy. Ecotoxicol Environ Saf 2019;167:178-87. [Crossref] [PubMed]
- Araya J, Tsubouchi K, Sato N, et al. PRKN-regulated mitophagy and cellular senescence during COPD pathogenesis. Autophagy 2019;15:510-26. [Crossref] [PubMed]
- Hara H, Kuwano K, Araya J. Mitochondrial Quality Control in COPD and IPF. Cells 2018;7:86. [Crossref] [PubMed]
- Nam HS, Izumchenko E, Dasgupta S, et al. Mitochondria in chronic obstructive pulmonary disease and lung cancer: where are we now? Biomark Med 2017;11:475-89. [Crossref] [PubMed]
- Mizumura K, Cloonan SM, Nakahira K, et al. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J Clin Invest 2014;124:3987-4003. [Crossref] [PubMed]
- Ito S, Araya J, Kurita Y, et al. PARK2-mediated mitophagy is involved in regulation of HBEC senescence in COPD pathogenesis. Autophagy 2015;11:547-59. [Crossref] [PubMed]
- Jiang S, Sun J, Mohammadtursun N, et al. Dual role of autophagy/mitophagy in chronic obstructive pulmonary disease. Pulm Pharmacol Ther 2019;56:116-25. [Crossref] [PubMed]
- Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223-7. [Crossref] [PubMed]
- Chen Y, Li Z, Chen X, Zhang S. Long non-coding RNAs: From disease code to drug role. Acta Pharm Sin B 2021;11:340-54. [Crossref] [PubMed]
- Soni DK, Biswas R. Role of Non-Coding RNAs in Post-Transcriptional Regulation of Lung Diseases. Front Genet 2021;12:767348. [Crossref] [PubMed]
- Bi H, Zhou J, Wu D, et al. Microarray analysis of long non-coding RNAs in COPD lung tissue. Inflamm Res 2015;64:119-26. [Crossref] [PubMed]
- Zhang P, Cao L, Zhou R, et al. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun 2019;10:1495. [Crossref] [PubMed]
- Prinz F, Kapeller A, Pichler M, et al. The Implications of the Long Non-Coding RNA NEAT1 in Non-Cancerous Diseases. Int J Mol Sci 2019;20:627. [Crossref] [PubMed]
- Chen C, Feng Y, Wang X. LncRNA ZEB1-AS1 expression in cancer prognosis: Review and meta-analysis. Clin Chim Acta 2018;484:265-71. [Crossref] [PubMed]
- Ming X, Duan W, Yi W. Long non-coding RNA NEAT1 predicts elevated chronic obstructive pulmonary disease (COPD) susceptibility and acute exacerbation risk, and correlates with higher disease severity, inflammation, and lower miR-193a in COPD patients. Int J Clin Exp Pathol 2019;12:2837-48. [PubMed]
- Ijiri N, Asai K, Tohda M, et al. lncRNA NEAT1 regulates IL-6 and IL-8 mRNA expression in lung epithelial cells stimulated by cigarette smoke. Respirology 2019;24:160. [Crossref]
- Wang Y, Hu SB, Wang MR, et al. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat Cell Biol 2018;20:1145-58. [Crossref] [PubMed]
- Yan W, Chen ZY, Chen JQ, et al. LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson's disease through stabilizing PINK1 protein. Biochem Biophys Res Commun 2018;496:1019-24. [Crossref] [PubMed]
- Huang Z, Zhao J, Wang W, et al. Depletion of LncRNA NEAT1 Rescues Mitochondrial Dysfunction Through NEDD4L-Dependent PINK1 Degradation in Animal Models of Alzheimer's Disease. Front Cell Neurosci 2020;14:28. [Crossref] [PubMed]
- Bi H, Wang G, Li Z, et al. Long Noncoding RNA (lncRNA) Maternally Expressed Gene 3 (MEG3) Participates in Chronic Obstructive Pulmonary Disease through Regulating Human Pulmonary Microvascular Endothelial Cell Apoptosis. Med Sci Monit 2020;26:e920793. [PubMed]
- Lin H, Zhang X, Feng N, et al. LncRNA LCPAT1 Mediates Smoking/ Particulate Matter 2.5-Induced Cell Autophagy and Epithelial-Mesenchymal Transition in Lung Cancer Cells via RCC2. Cell Physiol Biochem 2018;47:1244-58. [Crossref] [PubMed]
- Baginski TK, Dabbagh K, Satjawatcharaphong C, et al. Cigarette smoke synergistically enhances respiratory mucin induction by proinflammatory stimuli. Am J Respir Cell Mol Biol 2006;35:165-74. [Crossref] [PubMed]
- Jones B, Donovan C, Liu G, et al. Animal models of COPD: What do they tell us? Respirology 2017;22:21-32. [Crossref] [PubMed]
- Tang N, Dong Y, Liu J, et al. Silencing of Long Non-coding RNA NEAT1 Upregulates miR-195a to Attenuate Intervertebral Disk Degeneration via the BAX/BAK Pathway. Front Mol Biosci 2020;7:147. [Crossref] [PubMed]
- Li JW, Ren SH, Ren JR, et al. Nimodipine Improves Cognitive Impairment After Subarachnoid Hemorrhage in Rats Through IncRNA NEAT1/miR-27a/MAPT Axis. Drug Des Devel Ther 2020;14:2295-306. [Crossref] [PubMed]
- Caramori G, Casolari P, Di Gregorio C, et al. MUC5AC expression is increased in bronchial submucosal glands of stable COPD patients. Histopathology 2009;55:321-31. [Crossref] [PubMed]
- Tsubouchi K, Araya J, Kuwano K. PINK1-PARK2-mediated mitophagy in COPD and IPF pathogeneses. Inflamm Regen 2018;38:18. [Crossref] [PubMed]
- Liang F, Xiao Q, Huang K, et al. The 17-y spatiotemporal trend of PM2.5 and its mortality burden in China. Proc Natl Acad Sci U S A 2020;117:25601-8. [Crossref] [PubMed]
- Rajnoveanu AG, Rajnoveanu RM, Motoc NS, et al. COPD in Firefighters: A Specific Event-Related Condition Rather than a Common Occupational Respiratory Disorder. Medicina (Kaunas) 2022;2022, 58:239.
- Xing YF, Xu YH, Shi MH, et al. The impact of PM2.5 on the human respiratory system. J Thorac Dis 2016;8:E69-74. [PubMed]
- Song C, He J, Wu L, et al. Health burden attributable to ambient PM2.5 in China. Environ Pollut 2017;223:575-86. [Crossref] [PubMed]
- Hikichi M, Mizumura K, Maruoka S, et al. Pathogenesis of chronic obstructive pulmonary disease (COPD) induced by cigarette smoke. J Thorac Dis 2019;11:S2129-40. [Crossref] [PubMed]
- Cantor JO, Turino GM. COPD Pathogenesis: Finding the Common in the Complex. Chest 2019;155:266-71. [Crossref] [PubMed]
- Ju Y, Ma X, Li H, et al. Relationship between Air Pollution and Hospital Admissions for Chronic Obstructive Pulmonary Disease in Changchun, China: A Season-Stratified Case-Cross Study. Can Respir J 2021;2021:3240785. [Crossref] [PubMed]
- Jin SM, Youle RJ. PINK1- and Parkin-mediated mitophagy at a glance. J Cell Sci 2012;125:795-9. [Crossref] [PubMed]
- Kawajiri S, Saiki S, Sato S, et al. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett 2010;584:1073-9. [Crossref] [PubMed]
- Lazarou M, Sliter DA, Kane LA, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015;524:309-14. [Crossref] [PubMed]
- Yamada T, Dawson TM, Yanagawa T, et al. SQSTM1/p62 promotes mitochondrial ubiquitination independently of PINK1 and PRKN/parkin in mitophagy. Autophagy 2019;15:2012-8. [Crossref] [PubMed]
- Ahmad T, Sundar IK, Lerner CA, et al. Impaired mitophagy leads to cigarette smoke stress-induced cellular senescence: implications for chronic obstructive pulmonary disease. FASEB J 2015;29:2912-29. [Crossref] [PubMed]
- Mannam P, Rauniyar N, Lam TT, et al. MKK3 influences mitophagy and is involved in cigarette smoke-induced inflammation. Free Radic Biol Med 2016;101:102-15. [Crossref] [PubMed]
- Zhang HH, Li Z, Liu Y, et al. Physical and chemical characteristics of PM2.5 and its toxicity to human bronchial cells BEAS-2B in the winter and summer. J Zhejiang Univ Sci B 2018;19:317-26. [Crossref] [PubMed]
- Liu S, Liu M, Dong L. The clinical value of lncRNA MALAT1 and its targets miR-125b, miR-133, miR-146a, and miR-203 for predicting disease progression in chronic obstructive pulmonary disease patients. J Clin Lab Anal 2020;34:e23410. [Crossref] [PubMed]
- Song J, Wang Q, Zong L. LncRNA MIR155HG contributes to smoke-related chronic obstructive pulmonary disease by targeting miR-128-5p/BRD4 axis. Biosci Rep 2020;40:BSR20192567. [Crossref] [PubMed]
- Wright JL, Churg A. Animal models of cigarette smoke-induced COPD. Chest 2002;122:301S-6S. [Crossref] [PubMed]
- Zhang M, Shi R, Zhang Y, et al. Nix/BNIP3L-dependent mitophagy accounts for airway epithelial cell injury induced by cigarette smoke. J Cell Physiol 2019;234:14210-20. [Crossref] [PubMed]
- Willis-Owen SAG, Thompson A, Kemp PR, et al. COPD is accompanied by co-ordinated transcriptional perturbation in the quadriceps affecting the mitochondria and extracellular matrix. Sci Rep 2018;8:12165. [Crossref] [PubMed]


