
CHD7 in oocytes is essential for female fertility
Introduction
The chromodomain helicase DNA-binding protein 7 (CHD7) is an adenosine triphosphate (ATP)-dependent chromatin remodeler that plays pivotal functions during various developmental processes. ATP-dependent chromatin remodeling complexes consist of 4 subfamilies: the switching/sucrose nonfermenting (SWI-SNF) family, the imitation switch (ISWI) family, the CHD family, and the inositol-requiring 80 (INO80) family (1). These chromatin remodeling complexes share a similar ATPase-containing domain which can use ATP hydrolysis to alter histone-DNA contacts, exerting critical roles in normal development and cancer (2).
Previous studies have revealed that chromatin remodeling regulated by CHD7 is involved in key developmental processes at the transcriptional level, including neural crest cell formation, neuronal differentiation, great vessel, and cardiovascular development (3-6). CHD7 can regulate the transcription initiation and elongation at the enhancer and promoter region of the gene. For example, CHD7 can regulate neural differentiation in brain development by its transcriptional regulation in progenitor cells (7). Even heterozygous mutations of CHD7 can cause a severe congenital disease known as CHARGE (Coloboma, Heart defect, Atresia choanae, Restricted growth, Genital hypoplasia and Ear abnormality) syndrome, with pathogenic mutations at the rate of approximately 60% to 70%. CHARGE syndrome is a rare disorder characterized by a specific pattern of defects, including ocular coloboma, heart malformations, atresia of the nasal choanae, growth retardation, genital hypoplasia, and ear abnormalities (8). Although CHD7-related CHARGE syndrome manifests in genital abnormalities, including cryptorchidism, micropenis, and abnormal morphology of female internal genitalia, its physiological roles in the female reproductive system remain less understood (9,10). Many researchers attribute these symptoms to a deficiency of gonadotropin-releasing hormone (GnRH)-induced gonadotropin secretion, which is caused by decreased GnRH neurons in the hypothalamus and decreased GnRH receptors in the pituitary gland (11). In patients with congenital hypogonadotropic hypogonadism (CHH), the percentage of CHD7 variants has been reported to be approximately 6–16% (12,13). CHH is also characterized by a deficiency in GnRH-induced gonadotropin secretions (14). These gonadotropin deficiencies can result in delayed puberty and agenesis of the uterus and ovaries (15).
However, the clinical and molecular genetic features of patients with CHD7 allelic variants remain unclear. Firstly, not all patients with CHD7 alterations and CHH symptoms respond well to cyclical gonadotropin therapy (16). In addition, patients with CHH have an almost 10% spontaneous recovery of their hypothalamic-pituitary-gonadal (HPG) axis function in later adulthood (17,18). So far, no recovery of the HPG axis has ever been reported in female patients with CHD7 alterations and CHARGE, or in patients with CHD7 alterations and CHH, indicating the complexity of the functions of CHD7 in the female reproductive system. Jolly et al. performed whole-exome sequencing analysis and reported that CHD7 was related to primary ovarian insufficiency (POI) (19). However, the role of CHD7 in ovarian function is not fully understood. To date, apart from studies on the effects on GnRH neurons, no reports have investigated the specific role of CHD7 in the reproductive system, especially during oogenesis.
Previous evidence from single-cell RNA-seq data has shown high expression levels of CHD7 during human oogenesis (20). In the present study, we examined a mouse strain of Chd7-GFP transgenic mice. In these mice, we observed high expression levels of Chd7-GFP in oocytes at different stages, from newborn to adult, which indicated that CHD7 might have a specific function in the ovary. To our knowledge, this is the first study to explore the function of CHD7 in follicle development. Due to the lethality of systemic Chd7 knockout in mice, we could only conditionally delete Chd7 in mice follicles using the Cre-loxP approach with the transgenic mice carrying growth differentiation factor 9 (GDF9) promoter-mediated Cre recombinase. This precise Chd7 deletion in oocytes enabled us to explore the specific role of CHD7 during oogenesis, which could contribute to a better understanding of CHD7 function in the developmental stages of the female reproductive systems. We applied several important indicators to evaluate the ovary function like ovary size, fertility function, follicle counting. Small ovary size can give us the impression of the impaired ovary function and decreased ovarian reserve. Overall, we observed that conditional deletion of Chd7 led to reduced ovary size, decreased reproductive ability, decreased follicle counting numbers. Furthermore, deletion of Chd7 led to granulosa cell (GC) apoptosis during ovarian development, which is essential for normal oocyte formation. We presented the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-609/rc).
Methods
mRNA expression profiling
The online website Gene Expression Omnibus (GEO) was used to explore the mRNA expression profiling of CHD7 (https://www.ncbi.nlm.nih.gov/geo/). In this study, we analyzed the CHD7 expression at different stages of human follicle development in GSE107746 and Chd7 expression at different age of mouse germinal vesicle (GV) oocytes at 2, 9, and 14 months old from GSE159281. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Mice and genotyping
A protocol was prepared before the study without registration. All experiments were performed with the approval [No. 2019(135)] granted by the ethics committee of Children’s Hospital of Fudan University, in compliance with the Guidelines for the Ethical Review of Animal Welfare and the Guide for the Care and Use of Laboratory Animals for the care and use of animals. All mice (n=40) had a C57BL/6J genetic background. Chd7flox/flox(Chd7f/f) mice, Chd7-GFP transgenic mice, and Gdf9-Cre mice were obtained as previously described (21,22). Healthy female C57BL/6J mice were maintained as controls. All these mice were kept in pathogen-free conditions in an environment of 20±2 ℃ room temperature and 50–70% humidity, on a 12:12-hour light-dark cycle, with enough food and water. In the Gdf9-Cre mice, the Cre recombinase was specifically expressed in oocytes from primordial through to later stage follicles. As CHD7 is expressed in primordial follicle to mature oocytes, we used a Gdf9-Cre LoxP system to delete CHD7 in mouse oocytes. By crossing Chd7f/f mice with transgenic mice expressing GDF9 promoter-mediated Cre recombinase, we generated mice with no Chd7 expression in their oocytes. Mouse genotyping was carried out using the universal mouse genotyping protocol adapted from Stratman and Simon (23). The following primers were used to detect the Cre allele: 5'-ACCAGGTTCGTTCACTCATGG-3' (forward) and 5'-AGGCTAAGTGC CTTCTCTACA'-3 (reverse). To detect CHD7 flox band, the following primers were used: 5'-TGCAGATGGGACGTTTTCAG-3' (forward) and 5'-CTGCAAGAACACAGGGCAAG-3' (reverse).
Fertility assessment and ovarian histology analysis
To assess fertility, 8-week-old Gdf9-Cre:Chd7f/f and Chd7f/f female mice were mated with normal 10-week-old wild-type (WT) males for 6 months (2 females: 1 male) (n=4 for each group). The number and size of the offspring from each pregnant female mouse were recorded at embryonic day 19 (E19) or after birth. For histological analysis, the ovaries were fixed in 4% paraformaldehyde for 24 hours at room temperature. After being embedded in paraffin, tissues were cut into 5–6 µm serial sections and stained with hematoxylin and eosin (H&E). The remaining sections were stored at room temperature for later use.
Follicle counting
For follicle counting, every follicle with an oocyte at a different developmental stage was counted in every fifth section of the ovaries. Only oocytes with a clear visible nucleus were counted, and follicle counts at 4 stages were recorded as means ± standard deviation (SD) per ovary, per section. Follicle classifications were defined as previously described (24). Primordial follicles were defined as those with oocytes surrounded by only a single layer of squamous GCs. Primary follicles were defined as those with oocytes surrounded by at least 3 cuboidal GCs in a single layer. Secondary follicles were defined as those with 2 or more layers of GCs around the oocyte but no antral space in the granulosa layer. Antral follicles were defined as those with at least 2 layers of GCs around the oocyte with an antral space. Follicle counting was performed blindly by two experienced analysts to reduce bias.
Real-time quantitative polymerase chain reaction (RT-qPCR)
TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Carlsbad, USA) was used to extract total RNA from ovaries in the different groups according to the standard protocol. cDNAs were synthesized using a PrimeScript RT-PCR kit (Takara Bio, Shiga, Japan). RT-qPCR was conducted on an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, USA) using a SYBR Premix Ex Taq II kit (Takara Bio). The primer sequences for Chd7 were as follows: 5'-CAAAGCAGGGCCAGAACAAG-3' (forward), 5'-TCCCACGTGCTGTCTTCATA-3' (reverse). GAPDH was used as an internal control for mRNAs with primers as follows: 5'-CCCATCACCATCTTCCAGGAG-3' (forward), 5'-CTTCTCCATGGTGGTGAAGACG-3' (reverse). Gene expression was analyzed using the ΔΔCq method.
Immunohistochemistry (IHC) assay
After slides were deparaffinized in xylene, tissue sections were rehydrated. This was followed by antigen retrieval. After washing in phosphate-buffered saline (PBS), tissue sections were labeled with antibodies against CHD7 (1:100, HPA053075, Sigma, St. Louis, USA), GFP (1:3,200, ab6556, Abcam, Cambridge, UK), cleaved Caspase-3 (1:200, 9661, Cell Signaling Technology, Danvers, MA, USA), and Ki67 (1:200, ab15580, Abcam). Slides were observed and photographed recorded under a microscope (400×) (Olympus, Tokyo, Japan).
Immunofluorescence (IF) assay
The removed ovaries were fixed in 4% paraformaldehyde and then immersed into 30% sucrose in PBS. After the ovaries were embedded in optimum cutting temperature (OCT) compound at −78 ℃, they were serially sectioned into 15 µm thicknesses in a cryostat (Leica, Wetzlar, Germany) for later use. After the slides were washed with PBS and blocked with 5% normal serum, the primary antibodies (GFP: 1:3,200, ab6556, Abcam; Ki67 1:200, ab15580, Abcam) were incubated at 4 degrees overnight. Expression was observed instantly using a Leica DMIRB Inverted Fluorescence Microscope (Leica, Wetzlar, Germany). The nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI) 1:5,000 (Solarbio, Beijing, China).
Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay
A TUNEL assay was performed on the ovary paraffin sections using an in situ cell death detection kit (11684795910, Roche, Basel, Switzerland) according to the manufacturer’s instructions. The fluorescence signal was detected using a Leica DMIRB Inverted Fluorescence Microscope.
Statistical analysis
Data were presented as mean ± SD. Differences between the two groups were analyzed with t-tests. Multiple comparisons were analyzed with one-way analysis of variance (ANOVA) using GraphPad Prism version 8.0 (GraphPad Software, San Diego, USA). A P value <0.05 was considered statistically significant.
Results
High expression levels of CHD7 in human and mouse oocytes
Firstly, using data from the GEO database, we investigated the expression level of CHD7 during human folliculogenesis according to the gene expression profiles of GSE107746 (20). Single-cell RNA-seq revealed that CHD7 has a relatively high expression in oocytes from the primordial to the preovulatory stage, with especially high expression at the preovulatory follicle stage (Figure 1A). In contrast, the contemporary surrounding GCs had much lower CHD7 expression levels. In another single-cell RNA-seq analysis evaluating the expression levels of Chd7 in mouse oocytes and the surrounding cumulus at different ages, we obtained data showing high Chd7 expression levels in mouse GV oocytes at 2, 9, and 14 months old from GSE159281 (Figure 1B) (25).
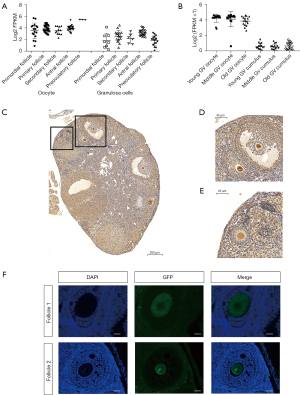
To evaluate Chd7 expression during mouse folliculogenesis, we examined a Chd7-GFP transgenic mouse strain. Using IHC, we observed that Chd7 had high expression from the primordial to preovulatory follicle stages in Chd7-GFP transgenic mice. (Figure 1C-1E). The expression pattern was similar in the IF results (Figure 1F). Overall, CHD7 had high expression levels in follicles at all stages but not in GCs. These results indicated that CHD7 might have a specific function in folliculogenesis.
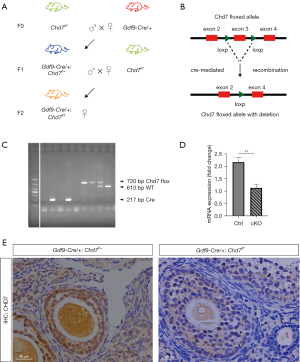
Generation of Gdf9-Cre:Chd7f/f conditional knockout (cKO) mice
By crossing Chd7f/fmice with transgenic mice expressing GDF9 promoter-mediated Cre recombinase, we generated Gdf9-Cre:Chd7f/f cKO mice (Figure 2A,2B). In Gdf9-Cre mice, Cre was specifically expressed in oocytes in primordial and later-stage follicles from postnatal day 21 (26). In the present study, the PCR product sizes for genotyping were 610 bp (WT) and 750 bp (Chd7 flox allele) (Figure 2C). We used IHC to confirm the deletion of Chd7 expression in the ovaries. Our results showed that CHD7 expression decreased in oocytes, confirming the successful generation of Chd7 oocyte-specific cKO mice (Figure 2D). Total RNA was extracted from the whole ovary for RT-qPCR, which revealed a significant decrease in Chd7 mRNA expression in the cKO mice compared to the control mice (Figure 2E).
Reduced fertility and decreased follicular numbers in cKO mice
We found that the cKO and WT mice had similar body sizes and weights at the age of 8 weeks. These cKO mice also showed normal behavior like WT mice without any abnormal phenotype. However, we observed that the cKO mice had significantly smaller ovaries and lower ovary weights compared to the WT mice (P<0.001) (Figure 3A). To test whether the deletion of Chd7 had effects on female fertility, we performed a breeding test by mating female cKO and control mice with WT males for 6 months. We found that the number of litters was markedly reduced in the cKO mice compared to the control (WT or heterozygous genotype) females (Figure 3B,3C). The cKO female mice were completely infertile or sub-infertile, suggesting that CHD7 is essential for female fertility.
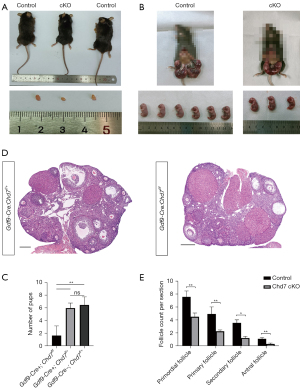
To further investigate possible defects in follicular development, we performed follicle counting in H&E-stained sections of the ovaries from control and cKO females. The H&E staining results allowed us to identify follicles at different stages in both genotypic groups (Figure 3D), and the follicle counting results revealed that the number of follicles at all 4 stages was markedly reduced in the cKO mice (P<0.01 in primordial, primary, and antral follicles, and P<0.05 in secondary follicles, n=3 for both groups) (Figure 3E).
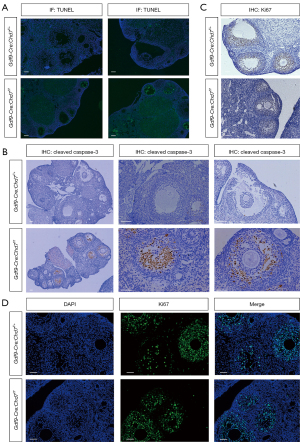
CHD7 deficiency promoted GC apoptosis and did not inhibit cell proliferation
As ovary size was remarkably smaller in the cKO mice, we investigated whether CHD7 would affect apoptosis and proliferation levels in the ovaries. We performed a TUNEL assay and confirmed that the apoptosis rate was much higher in the GCs of the cKO mice than in the control mice, most notably in the preantral and early antral follicles (Figure 4A). The percentage of atretic follicles was significantly greater in the cKO mice than in the control mice. IHC indicated that the expression of cleaved caspase-3 positive cells in the cKO group was significantly higher than in the control group (Figure 4B). In addition, we used Ki-67 staining to detect cell proliferation in the cKO and WT mouse ovaries. IHC and IF results showed no difference between the cKO and control groups (Figure 4C,4D).
Discussion
ATP-dependent chromatin remodelers play critical roles in follicular development. These family members can change the chromatin structure by repositioning, mobilizing, and restructuring nucleosomes, using energy from ATP hydrolysis (27). Previous studies have confirmed that these family members have diverse functions during oogenesis. In 2006, Bultman et al. reported that depleting maternal BRG1 after oocyte development resulted in 2-cell arrest, thus revealing BRG1’s regulatory function in zygotic genome activation (ZGA) in mice (28). Zhang et al. also reported that SNF2h (also known as Smarca5) plays a crucial role in oocyte maturation in mouse oocytes, showing that the loss of SNF2h could result in the dysregulation of several genes involved in maturation-promoting factor (MPF) activation (29). Another chromatin-remodeling factor, SNF2l (also known as Smarca1), has been shown to be required for oocyte superovulation and capable of regulating FGL2 in differentiating mouse GCs (30).
The CHD family includes the CHDs 1–9, which can regulate transcriptional activation and DNA repair (31). An important member of the CHD family, CHD7, is notorious for causing the recognizable genetic CHARGE syndrome. In addition, CHD7 mutations are also associated with idiopathic hypogonadotropic hypogonadism (IHH) and Kallmann syndrome (32). CHD7-related reproductive phenotypes vary widely, from primary amenorrhea to delayed puberty and infertility. Previous studies have attributed these symptoms to a deficiency of hypothalamic GnRH (33,34). In recent years, as the understanding of the clinical features and molecular mechanisms of these CHD7 alterations has improved, more researchers have realized the complexities of CHD7 function in the reproductive system. To date, most studies have not considered the specific function of CHD7 in folliculogenesis, independent of GnRH.
Our study attempted to determine the specific function of CHD7 in folliculogenesis. We first confirmed the high expression levels of CHD7 in mouse ovaries. Using Chd7-GFP transgenic mouse, we identified the expression patterns of CHD7 in the ovaries. With the creation of an approximately 200 kb-long bacterial artificial chromosome (BAC) containing the Chd7 gene, we found that the Chd7-GFP mice analogized the expression pattern of the CHD7 gene in their ovaries. The BAC address for this line was RP23-127L15, which overlapped with the promoter and coding sequences of Chd7.
Transgenic mice, Gdf9-Cre and Zp3-Cre, are two common mice which allows inactivation of a target gene at different points during folliculogenesis. In the GDF9-Cre mice, the Cre recombinase was specifically expressed in oocytes from primordial through to later stage follicles (35). In Zp3-Cre mice, the Cre gene is driven by the zona pellucida protein 3 (Zp3) promoter which express the Cre recombinase in the oocytes at primary and/or later follicular stage (36). As CHD7 is expressed in primordial through to mature oocytes, we used a Gdf9-Cre LoxP system to delete CHD7 in mouse oocytes. After successfully generating the Gdf9-Cre:Chd7f/f mice, we were able to further analyze the function of CHD7 during oogenesis.
To our knowledge, our study was the first to observe Chd7-cKO mice showing infertility or sub-fertility, smaller ovary sizes, decreased preovulatory follicle numbers, and increased follicular atresia. These findings suggest that CHD7 could have a specific and important function in follicular development. In addition, we observed significantly higher GC apoptosis rates in the preovulatory follicles of cKO mice. The bidirectional interactions between oocytes and the GCs surrounding them are important for oocyte maturation because many steroidal hormones, like luteinizing hormone (LH) and follicle-stimulating hormone (FSH), and growth factors, such as insulin-like growth factor 1 (IGF-1) and epidermal growth factor (EGF) can benefit oocyte maturation and prevent cell apoptosis in preovulatory follicles (37,38). As a synchronous partnership, the relationship between the oocyte and its supporting GCs is vital (39). Oocytes can direct GC proliferation, differentiation, and apoptosis, while GCs can influence oocyte maturation.
Our preliminary results showed significant differences in apoptosis but not in proliferation of GCs between the cKO and control groups. The balance between pro-apoptotic and anti-apoptotic factors in GCs has been shown to be vital during folliculogenesis (40). Apoptosis of GCs can mediate follicular atresia, and our study found that greater follicular atresia in oocyte which is related with high apoptosis rates in Chd7-deficient mouse oocytes. Although the development of ovarian follicles is precisely co-regulated by pituitary-induced gonadotropins together with local paracrine factors in the ovary, the present study confirmed that CHD7 could exert a unique function in oocyte development by targeting the oocyte directly. Thus, we should consider that patients with CHD7 alterations may not respond well to GnRH treatment.
This study had some limitations. First, we did not analyze the specific defects of these oocytes in the cKO mice. Secondly, as a chromatin remodeling factor, CHD7 regulates the transcription of many downstream genes through a process known as chromatin remodeling. More experiments are needed to explore the detailed mechanisms underlying this phenomenon.
Conclusions
To our knowledge, our study has been the first to confirm that CHD7 plays a crucial role in suppressing follicular atresia and promoting oocyte maturation.
Acknowledgments
Funding: This work was supported by the Shanghai Sailing Program (No. 20YF1425100), the National Natural Science Foundation of China (No. 81974229, for W Feng).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-609/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-609/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-609/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All experiments were performed with the approval [No. 2019(135)] granted by the ethics committee of Children’s Hospital of Fudan University, in compliance with the Guidelines for the Ethical Review of Animal Welfare and the Guide for the Care and Use of Laboratory Animals for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 2011;21:396-420. [Crossref] [PubMed]
- Li Y, Gong H, Wang P, et al. The emerging role of ISWI chromatin remodeling complexes in cancer. J Exp Clin Cancer Res 2021;40:346. [Crossref] [PubMed]
- Bajpai R, Chen DA, Rada-Iglesias A, et al. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 2010;463:958-62. [Crossref] [PubMed]
- Feng W, Kawauchi D, Körkel-Qu H, et al. Chd7 is indispensable for mammalian brain development through activation of a neuronal differentiation programme. Nat Commun 2017;8:14758. [Crossref] [PubMed]
- Randall V, McCue K, Roberts C, et al. Great vessel development requires biallelic expression of Chd7 and Tbx1 in pharyngeal ectoderm in mice. J Clin Invest 2009;119:3301-10. [Crossref] [PubMed]
- Yan S, Thienthanasit R, Chen D, et al. CHD7 regulates cardiovascular development through ATP-dependent and -independent activities. Proc Natl Acad Sci U S A 2020;117:28847-58. [Crossref] [PubMed]
- Feng W, Shao C, Liu HK. Versatile Roles of the Chromatin Remodeler CHD7 during Brain Development and Disease. Front Mol Neurosci 2017;10:309. [Crossref] [PubMed]
- Sanlaville D, Verloes A. CHARGE syndrome: an update. Eur J Hum Genet 2007;15:389-99. [Crossref] [PubMed]
- Umino S, Kitamura M, Katoh-Fukui Y, et al. A case of combined 21-hydroxylase deficiency and CHARGE syndrome featuring micropenis and cryptorchidism. Mol Genet Genomic Med 2019;7:e730. [Crossref] [PubMed]
- Tellier AL, Cormier-Daire V, Abadie V, et al. CHARGE syndrome: report of 47 cases and review. Am J Med Genet 1998;76:402-9. [Crossref] [PubMed]
- Layman WS, Hurd EA, Martin DM. Reproductive dysfunction and decreased GnRH neurogenesis in a mouse model of CHARGE syndrome. Hum Mol Genet 2011;20:3138-50. [Crossref] [PubMed]
- Gonçalves CI, Patriarca FM, Aragüés JM, et al. High frequency of CHD7 mutations in congenital hypogonadotropic hypogonadism. Sci Rep 2019;9:1597. [Crossref] [PubMed]
- Boehm U, Bouloux PM, Dattani MT, et al. Expert consensus document: European Consensus Statement on congenital hypogonadotropic hypogonadism--pathogenesis, diagnosis and treatment. Nat Rev Endocrinol 2015;11:547-64. [Crossref] [PubMed]
- Balasubramanian R, Crowley WF Jr. Reproductive endocrine phenotypes relating to CHD7 mutations in humans. Am J Med Genet C Semin Med Genet 2017;175:507-15. [Crossref] [PubMed]
- Reynaert N, de Zegher F, Francois I, et al. Expanding the CHARGE Geno-Phenotype: A Girl with Novel CHD7 Deletion, Hypogonadotropic Hypogonadism, and Agenesis of Uterus and Ovaries. Horm Res Paediatr 2016;85:288-90. [Crossref] [PubMed]
- Zhang L, Gao Y, Du Q, et al. Genetic Profiles and Three-year Follow-up Study of Chinese Males With Congenital Hypogonadotropic Hypogonadism. J Sex Med 2021;18:1500-10. [Crossref] [PubMed]
- Raivio T, Falardeau J, Dwyer A, et al. Reversal of idiopathic hypogonadotropic hypogonadism. N Engl J Med 2007;357:863-73. [Crossref] [PubMed]
- Sidhoum VF, Chan YM, Lippincott MF, et al. Reversal and relapse of hypogonadotropic hypogonadism: resilience and fragility of the reproductive neuroendocrine system. J Clin Endocrinol Metab 2014;99:861-70. [Crossref] [PubMed]
- Jolly A, Bayram Y, Turan S, et al. Exome Sequencing of a Primary Ovarian Insufficiency Cohort Reveals Common Molecular Etiologies for a Spectrum of Disease. J Clin Endocrinol Metab 2019;104:3049-67. [Crossref] [PubMed]
- Zhang Y, Yan Z, Qin Q, et al. Transcriptome Landscape of Human Folliculogenesis Reveals Oocyte and Granulosa Cell Interactions. Mol Cell 2018;72:1021-1034.e4. [Crossref] [PubMed]
- Feng W, Khan MA, Bellvis P, et al. The chromatin remodeler CHD7 regulates adult neurogenesis via activation of SoxC transcription factors. Cell Stem Cell 2013;13:62-72. [Crossref] [PubMed]
- Sheng X, Liu C, Yan G, et al. The mitochondrial protease LONP1 maintains oocyte development and survival by suppressing nuclear translocation of AIFM1 in mammals. EBioMedicine 2022;75:103790. [Crossref] [PubMed]
- Stratman JL, Barnes WM, Simon TC. Universal PCR genotyping assay that achieves single copy sensitivity with any primer pair. Transgenic Res 2003;12:521-2. [Crossref] [PubMed]
- Myers M, Britt KL, Wreford NG, et al. Methods for quantifying follicular numbers within the mouse ovary. Reproduction 2004;127:569-80. [Crossref] [PubMed]
- Mishina T, Tabata N, Hayashi T, et al. Single-oocyte transcriptome analysis reveals aging-associated effects influenced by life stage and calorie restriction. Aging Cell 2021;20:e13428. [Crossref] [PubMed]
- Sun QY, Liu K, Kikuchi K. Oocyte-specific knockout: a novel in vivo approach for studying gene functions during folliculogenesis, oocyte maturation, fertilization, and embryogenesis. Biol Reprod 2008;79:1014-20. [Crossref] [PubMed]
- Smale ST, Fisher AG. Chromatin structure and gene regulation in the immune system. Annu Rev Immunol 2002;20:427-62. [Crossref] [PubMed]
- Bultman SJ, Gebuhr TC, Pan H, et al. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev 2006;20:1744-54. [Crossref] [PubMed]
- Zhang C, Chen Z, Yin Q, et al. The chromatin remodeler Snf2h is essential for oocyte meiotic cell cycle progression. Genes Dev 2020;34:166-78. [Crossref] [PubMed]
- Pépin D, Paradis F, Perez-Iratxeta C, et al. The imitation switch ATPase Snf2l is required for superovulation and regulates Fgl2 in differentiating mouse granulosa cells. Biol Reprod 2013;88:142. [Crossref] [PubMed]
- Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res 2007;618:30-40. [Crossref] [PubMed]
- Kim HG, Kurth I, Lan F, et al. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet 2008;83:511-9. [Crossref] [PubMed]
- Topaloğlu AK. Update on the Genetics of Idiopathic Hypogonadotropic Hypogonadism. J Clin Res Pediatr Endocrinol 2017;9:113-22. [PubMed]
- Xu C, Cassatella D, van der Sloot AM, et al. Evaluating CHARGE syndrome in congenital hypogonadotropic hypogonadism patients harboring CHD7 variants. Genet Med 2018;20:872-81. [Crossref] [PubMed]
- Lan ZJ, Xu X, Cooney AJ. Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol Reprod 2004;71:1469-74. [Crossref] [PubMed]
- Lewandoski M, Wassarman KM, Martin GR. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr Biol 1997;7:148-51. [Crossref] [PubMed]
- Hsieh M, Zamah AM, Conti M. Epidermal growth factor-like growth factors in the follicular fluid: role in oocyte development and maturation. Semin Reprod Med 2009;27:52-61. [Crossref] [PubMed]
- Glister C, Tannetta DS, Groome NP, et al. Interactions between follicle-stimulating hormone and growth factors in modulating secretion of steroids and inhibin-related peptides by nonluteinized bovine granulosa cells. Biol Reprod 2001;65:1020-8. [Crossref] [PubMed]
- Chang HM, Qiao J, Leung PC. Oocyte-somatic cell interactions in the human ovary-novel role of bone morphogenetic proteins and growth differentiation factors. Hum Reprod Update 2016;23:1-18. [Crossref] [PubMed]
- Regan SLP, Knight PG, Yovich JL, et al. Granulosa Cell Apoptosis in the Ovarian Follicle-A Changing View. Front Endocrinol (Lausanne) 2018;9:61. [Crossref] [PubMed]


