
Novel germline mutation in lung cancer pedigrees establishes BCAR1 as a human cancer susceptibility gene: a case report
Introduction
Lung cancer is the most prevalent malignancy worldwide, in which non-small cell lung cancer (NSCLC) is the predominant histologic type, mainly including adenocarcinoma and squamous cell carcinoma (1). Smoking is the primary non-inherited risk factor for lung cancer; however, approximately 25% of lung cancer cases worldwide are not attributable to tobacco use (2). Multiple risk factors, including environment, hormones, and genetic predisposition, have been associated with the pathogenesis of lung cancer in never smokers. Among these factors, familial lung cancer has been reported (3), and people with family history of lung cancer have double the risk of developing the disease (4,5). In the past decades, numerous somatic mutations have been identified in lung cancer tissues and contributed to the establishment of an oncogene-centric molecular classification paradigm in this disease. In contrast, relatively few germline mutations have been identified. Although familial forms of lung cancer are rarely described, several susceptibility genes have been identified, such as EGFR (3,6), HER2 (7), BAP1 (8), and BRCA2 (9), among which EGFR germline mutations have been the most reported.
In this report, we have presented 2 siblings with lung cancer harboring germline mutation in breast cancer anti-estrogen resistance protein 1 (BCAR1). Somatic mutations and germline mutations including BCAR1 and FAM20C were identified in the 2 siblings through whole exome sequencing (WES). Further frameshift germline mutation of BCAR1 was confirmed in the proband by Sanger sequencing, and screening across other family members revealed their presence in 2 affected individuals. To our knowledge, this is the first time that frame-shift germline mutations of BCAR1 were found by WES and Sanger sequencing in lung cancer patients. Furthermore, the BCAR1 gene, encoding a scaffold protein, has been shown to be overexpressed and linked to adverse features in lung cancer (10-12). Therefore, we suspected that BCAR1 germline mutation potentially predisposes patients to lung cancer. We present the following article in accordance with the CARE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-7017/rc).
Case presentation
Tumor tissues and blood from 2 patients were collected in 2018. According to the digestive system tumor classification of the World Health Organization (WHO) in 2010, tumor samples were identified by hematoxylin and eosin (H&E) stained slides. The tumor tissue samples were histologically confirmed as lung squamous cell carcinoma and lung adenocarcinoma respectively by 2 molecular pathologists, and the tumor cell content exceeded 70%. All procedures performed in this study were in accordance with the ethical standards of the institutional and or national research committee(s) and with the Helsinki Declaration (as revised in 2013) and ethical standards of the Medical Ethics Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology and got its approval (ethical approval No. S412). Written informed consent was obtained from both patients for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
We used WES to analyse the formalin-fixed paraffin-embedded (FFPE) tissues and blood samples of the 2 siblings. Somatic mutations were obtained from tumor tissues and blood leukocytes, and germline mutations were obtained from blood leukocytes. As for individual mutational differences, mutations can be divided into somatic mutations and germline mutations. Somatic mutations are analysed using WES data of tumor samples with comparison to paired normal tissue for each individual. Somatic mutations are tumour-specific mutations after filtering out germline mutations which can be observed in the paired normal control samples as well. Therefore, it reflects individual differences between tumors. Germline mutations are defined as a gene alteration found in both tumor and paired normal control samples. They are individual-specific mutations in healthy tissues with comparison to reference genomes, reflecting individual differences and gene polymorphism in the population. The DNA was isolated from tumor FFPE tissues and blood leukocytes of blood samples using the DNeasy Blood and Tissue Kit (69504, QIAGEN, Venlo, Netherlands) according to the manufacturer’s instructions. The DNA quantity was assessed by Agilent’s Bioanalyzer (Agilent Technology, Santa Clara, CA, USA). The targeted capture pulldown and exon-wide libraries from genomic DNA were generated through the TruePrep DNA Library Prep Kit V2 for Illumina (#TD501, Vazyme, Nanjing, China). The sequences of captured libraries were performed as pair-end reads on sequences on the Illumina HiSeq 2500 platform. Variants were called using the Genome Analysis ToolKit (GATK; https://gatk.broadinstitute.org/hc/en-us) and functional annotation was performed through Annovar (https://annovar.openbioinformatics.org/en/latest/) and data were filtered by public databases, such as ExAC, 1000 Genomes Project, and Huma Gene Mutation Database (HGMD).
The blood samples of the 2 siblings and the 17 family members (except for family II: 6, 8 and family III: 6) were analysed by Sanger sequencing. The DNA was isolated from blood leukocytes of blood samples, germline BCAR1 and FAM20C mutations of the 2 siblings were further confirmed by Sanger sequencing, and screening across all 17 family members by Sanger sequencing revealed their presence in family members. Resequencing of BCAR1 c.942delinsAATGC CAGGGC (NM_001170717) was performed using the primers 5'-AAGTCTGTGCCTCTGCTT-3' (forward) and 5'-TTCCTCATCTGTCAACCATC-3' (reverse), amplified by polymerase chain reaction (PCR) at an annealing temperature of 58 °C. Resequencing of FAM20C c.951delinsGGACAGGTGAGCCCTTCCTTCCTCCCTC CATCCGC (NM_020223) was performed using the primers 5'-CATATGAGGAAC CCAGCACGTC-3' (forward) and 5'-AAAGGTGACGATGACATACAGGA-3' (reverse), amplified by polymerase chain reaction at an annealing temperature of 60 °C. Multiple sequencing reactions were loaded onto multiwell plates, which were injected into capillaries for electrophoresis on the instrument. Fluorescent-labeled modified (dideoxy) nucleotides were used that terminate the formation of a new DNA strand as they encounter their complementary nucleotides in the target sequence. This resulted in DNA strands of variable length, which were separated on a gel by electrophoresis and reflected the sequence being analyzed. Each capillary was separately calibrated for the dyes used in the sequencing reactions so that the software could perform the multi-component analysis to identify each of the dye-labeled fragments. These sequencing reactions were then analyzed using Variant Reporter V1.1.
Patient 1
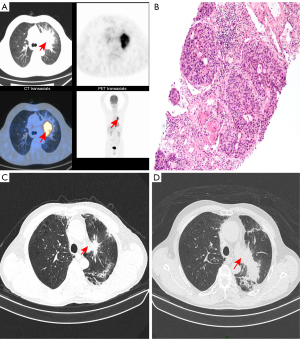
Patient 1, a 68-year-old male with smoking history, sought medical consultation for a 1-week history of cough and hemoptysis without any inducing factors, and was diagnosed with space-occupying lesions of the left upper lung in a local hospital. He was transferred to our department in May 2018 for further management. The chest computed tomography (CT) and positron emission tomography (PET)-CT confirmed a 6.4 cm × 5.2 cm mass in the left upper lung (Figure 1A), and multiple nodules were also found. A CT-guided lung biopsy revealed a NSCLC with a histological type of squamous cell (Figure 1B). The patient was diagnosed with stage IIIc lung squamous cell carcinoma (cT4N3M0) based on clinical-pathologic-radiographic correlation. Panel detection of cancer driver genes revealed 3 common somatic mutations associated with lung cancer including EGFR, PIK3CA, and TP53. He then underwent 2 cycles of chemotherapy using albumin paclitaxel and nedaplatin on 30 May 2018 and 22 June 2018, followed by radiotherapy (IMRT, PTV 60 Gy/30 F) on 12 July 2018. The subsequent CT revealed that the tumor had shrunk (Figure 1C). However, the patient was readmitted for cough, chest distress, and asthma in January 2019. The repeated CT scan revealed tumor recurrence (Figure 1D). The patient refused further treatment, and he died in July 2019.
Patient 2
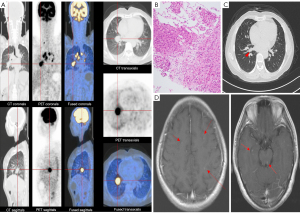
Patient 2, a 51-year-old female without a smoking history, was the younger sister of patient 1. She had experienced intermittent coughing for 1 month and sought medical consultation at a local hospital. Auxiliary examination revealed the presence of a nodule in the right parahilar region, as well as an increased number and enlarged size of mediastinal lymph nodes. She was transferred to our department in June 2018. Subsequent PET-CT and enhanced CT showed a 2.7 cm × 2.2 cm mass in the right lower lung (Figure 2A). Multiple enlarged lymph nodes were also identified in the right parahilar region and mediastinum. A CT-guided lung biopsy was performed on 12 June 2018. Pathological examination indicated lung adenocarcinoma (Figure 2B) with pleural metastasis, clinical stage was IVA (cT4N3M1a). The patient received 1 cycle of chemotherapy (pemetrexed disodium 800 mg/dL + carboplatin 0.5 g/dL) on 17 June 2018, and then started treatment with crizotinib. A follow-up CT scan demonstrated partial response with the indication of tumor shrinkage (Figure 2C). On 28 August 2019, lung lesions were stable; however, brain magnetic resonance imnaging (MRI) revealed potential metastasis (Figure 2D). The patient is currently under treatment with alectinib.
Discussion
With the development of next-generation sequencing (NGS) technology, gene mutations and dynamic epigenetic changes have further deepened our understanding of lung cancer and provided clues for new therapeutic targets. Molecular testing has become an integral part of lung cancer management. The detection of EGFR, BRAF and MET mutations as well as the analysis of ALK, ROS1, RET and NTRK translocations have already been incorporated into the diagnostic criteria for NSCLC (13). These oncogenic drivers strongly influence the formation and progression of lung cancer (6,14,15). Epigenetic regulation of genes is divided into different aspects, including DNA methylation, histone modifications and regulation of non-coding RNAs. Unlike direct gene mutations, epigenetic alterations of genes often lead to changes in intracellular RNA and protein expression levels leading to cancer. For example, aberrant DNA methylation promotes carcinogenesis through promoter methylation of tumor suppressor genes (TSG) to suppress their expression (16,17). The epigenetic disruption of tumor suppressor miRNA or oncomirs by promoter CpG methylation is common and can promote pro-tumoral capacities. Post-translational modifications of histones are also associated with lung carcinogenesis (18,19). Many studies have shown that key TSG or oncogenes are regulated by aberrant expression of miRNA, resulting in the acquisition of the characteristics of lung cancer (20-22). Overall, these data all emphasize the critical role of both gene mutations and multiple epigenetic events in the development of lung cancer.
In recent years, the developing technology of WES has shown enormous potential. This technology is now rapidly entering the clinical laboratories and changing the landscape of clinical testing and diagnostics, including to search for potential oncogenes to provide reference for molecular diagnosis of tumors (23,24). WES has achieved great success in identifying pathogenic variants for rare diseases and continued to contribute to the discovery of novel genes as well as the expansion of the phenotypic spectrum of known genes (25,26). The best drug targets related to tumor gene mutations can be obtained through WES technology, thereby changing the corresponding metabolic pathways, which may become the gold standard for individualized tumor therapy in the future (27,28). In addition, obtaining tumor gene mutations and DNA methylation information through WES technology can prompt tumor prognosis and guide clinical practice (29-31). Chang et al. applied an exome sequencing technology to determine variations and mutations in eight commonly used cancer cell lines, the results show that exome sequencing can be a reliable and cost effective way for identifying alterations in cancer genomes (32). Varela et al. applied WES on a series of primary clear cell renal cell carcinoma (ccRCC) (33). PBRM1, encoding for SWI/SNF chromatin remodeling complex was discovered in this study. There are also several studies that have attempted to identify driver mutations of cancer including leukemias, myelomas and solid tumors (34-36). In our report, WES was used to analyze the genetic characteristics of the two siblings.
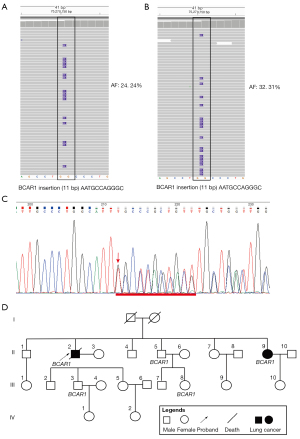
The results show that driver genes of EGFR, PIK3CA, and TP53 were identified in patient 1. Only a few distinct somatic mutations (patient 1 and patient 2, 21 and 4 somatic mutations, respectively) were identified in the 2 siblings (Table 1). In contrast, analysis of WES data from leukocytes revealed 479 germline mutations between the 2 siblings (https://cdn.amegroups.cn/static/public/10.21037atm-21-7017-1.xlsx). Surprisingly, 2 shared frameshift germline mutations (BCAR1 and FAM20C) were also identified by analyzing WES data from white blood cells of the 2 patients. We identified the frameshift germline mutations of BCAR1 in exon 4 (NM_001170717: c.942delinsAATGCCAGGGC) (Figure 3A,3B) and FAM20C in exon 4 (NM_020223: c.951delinsGGACAGGTGAGCCCTTCCTTCCTCCCTCCA TCCGC). In addition, the 2 siblings with BCAR1 and FAM20C variants were both identified as heterozygote mutations. Mutations in FAM20C cause an osteosclerotic bone dysplasia in humans known as Raine syndrome (37). However, another novel germline mutation of FAM20C was found in our report, encoding a novel frameshift germline mutation c.951delinsGGACAGGTGAGCCCTTCCTTCC TCCCTCCATCCGC, which showed a benign mutation (https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000402845.3). To further verify the hypothesis that germline mutations in BCAR1 may be associated with lung cancer susceptibility gene in patients, the mutation sites of the 2 genes were sequenced in the 2 siblings by Sanger sequencing. Both patients harbored a germline insertion of 11 bp (NM_001170 717: c.942delinsAATGCCAGGGC) producing a frameshift at codon 314 (Figure 3C). The BCAR1 mutation was confirmed by Sanger sequencing, and screening across all the 17 family members (except for family II: 6, 8 and family III: 6) revealed its presence in 2 affected individuals participating in the linkage study and in a further 3 relatives (Figure 3D). Several investigations have revealed that BCAR1 is an oncogene involved in metastasis and progression of multiple cancer types (38,39). Li et al. found that more deleterious BCAR1 mutations were detected in the HER2+ breast cancer subtype than in the HER2-breast cancer subtype, suggesting that BCAR1 mutations lead to its overactivation and are associated with ERBB2 overexpression (40), becoming resistant to the anti-proliferative effects of tamoxifen and leading to aggressive tumor progression (41-43). Overexpression of BCAR1 was associated with activation of p38 in NSCLC cases, and BCAR1 knockdown caused reduction of phospho-p38 levels in A549 cells (44). Multiple mutations in the BCAR1 protein have also been reported to contribute to ovarian cancer susceptibility (45). Huang et al. evaluated the predictive power of BCAR1 as a marker for poor prognosis in NSCLC cases (44). The results show that higher BCAR1 levels were strongly associated with more poorly differentiated NSCLC and predicted poorer prognosis. Multiple mutations of BCAR1 may affect its or different pathway-related genes expression levels, which may lead to different clinical syndromes. In order to clarify the clinical significance of BCAR1 expression in circulating tumor cells (CTCs) in the peripheral blood and tumor tissues in patients with early stage lung adenocarcinoma (ES-LUAD), Jiang et al. analyzed the predictive power of BCAR1 expression in CTCs and tumor tissues for disease-free survival (DFS). The results found that BCAR1 may have a “dual impact” on markers of epithelial-mesenchymal transition (EMT) in tumor tissues and CTCs due to micro-environmental disparities, resulting in important clinical significance, which can potentially guide accurate treatment of early stage lung adenocarcinoma (46). Frameshift germline mutation of BCAR1 is exceedingly rare, which is not presented in any online databases of human genetic variation, including the Exome Aggregation Consortium (ExAC) database, indicating it as a novel variant. In this study, frameshift germline mutation of BCAR1 was confirmed in the proband by Sanger sequencing, and screening across other family members revealed their presence in 2 affected individuals. The BCAR1 variant was identified in family II (Figure 3D), indicating that the allele frequency (AF) of BCAR1 variant was approximately 50% (heterozygote). The heterozygous BCAR1 mutation was identified in a heterozygous state in both patients. The BCAR1 gene encodes p130cas protein which possesses an amino-terminal Src-homology 3 domain, a substrate binding domain, proline-rich and serine-rich regions, and a bipartite Src-binding domain in carboxy-terminal sequence (10,47). The p130Cas protein itself has no intrinsic enzymatic or transcriptional activity, but they could control signaling events via phosphorylation and dephosphorylation and their association with effector proteins in multimolecular complexes. Tyrosine phosphorylation is the major post-translational modification of p130Cas, mainly occurring in the substrate binding domain, which helps to open binding sites for a variety of effector proteins (11). Many studies have demonstrated that p130Cas plays a vital role in cell migration (48,49), apoptosis (50,51), and cell transformation (52). The p130Cas protein has also been reported as involved in the development and progression of several human cancers, including breast cancer (53,54), prostate cancer (55,56), and lung cancer (44). In lung cancer, BCAR1 is usually overexpressed, and increased BCAR1 predicts poorer prognosis in NSCLC (44,57). High BCAR1 expression induced EMT in lung cancer cells to promote lung cancer cell migration and invasion (44,57). Mao et al. confirmed that the higher BCAR1-mRNA and protein expression levels in lung cancer were associated with poorer patient prognosis (58). The MTT assay and cell plate clone formation assay confirmed that BCAR1 plays an important role in cell proliferation, survival and clone formation in lung cancer. BCAR1 was confirmed to promote proliferation and cell growth by immunoprecipitation and mass spectrometry (IP-MS) assay, TCGA bioinformatics analysis and protein interaction (PPI-String) database, probably through upregulation of POLR2A and subsequent enhancement of catalase and transferase activities. In this report, BCAR1 variant occurred in 11 bp insertion at codon 314 which is located at the substrate binding domain containing 15 repeats of the YxxP consensus phosphorylation motif for Src family kinases (https://www.uniprot.org/uniprot/P56945#showFeaturesTable). The Src kinase binds p130Cas at its carboxy-terminal binding site, and is responsible for the direct phosphorylation of YxxP motifs in the substrate domain, which mediates the interaction of p130Cas with Crk adaptor proteins (10). The insertion of BCAR1 variant in the substrate binding domain indicates that its association with effector proteins would be impaired in some extent, thus leading to a wide range of effects, affecting different pathways, including cardiac development, endothelial migration, and cancer. The c.942delinsAATGC CAGGGC mutation is already presented in the BCAR1 gene, in 2 consecutive repeats, becoming triplicated in the proband family germline. The results indicate that this genetic region may be unstable and prone to duplication. Intriguingly, frameshifts often occur in repeated DNA sequences, and potential consequences of the 11 bp insertion in BCAR1 may lead to a truncated protein.
Table 1
| Sample | Chromosome | Position | Reference | Variant | Gene | Type | snp150Common | InterVar_automated | cytoBand |
|---|---|---|---|---|---|---|---|---|---|
| Patient 1 | chr1 | 78414310 | – | AA | FUBP1 | 1p31.1 | |||
| Patient 1 | chr1 | 120612040 | – | CCTCCGCCG | NOTCH2 | 1p11.2 | |||
| Patient 1 | chr2 | 112786523 | – | T | MERTK | rs147500027 | 2q13 | ||
| Patient 1 | chr3 | 189507518 | – | AGAG | TP63 | 3q28 | |||
| Patient 1 | chr5 | 56177849 | CAA | – | MAP3K1 | Nonframeshift deletion | rs5868032 | 5q11.2 | |
| Patient 1 | chr6 | 43738449 | – | GACA | VEGFA | Frameshift insertion | 6p21.1 | ||
| Patient 1 | chr6 | 157100431 | GGC | – | ARID1B | Nonframeshift deletion | 6q25.3 | ||
| Patient 1 | chr6 | 163991700 | – | T | QKI | rs555941441 | 6q26 | ||
| Patient 1 | chr7 | 151945071 | - | T | KMT2C | Stopgain | rs150073007 | 7q36.1 | |
| Patient 1 | chr8 | 145738768 | G | - | RECQL4 | 8q24.3 | |||
| Patient 1 | chr9 | 133759490 | AAG | - | ABL1 | Nonframeshift deletion | rs201725154 | 9q34.12 | |
| Patient 1 | chr11 | 32452241 | C | - | WT1 | rs76500597 | 11p13 | ||
| Patient 1 | chr12 | 121434630 | – | TCATTCAT | HNF1A | 12q24.31 | |||
| Patient 1 | chr13 | 28942678 | ATG | – | FLT1 | rs138306957 | 13q12.3 | ||
| Patient 1 | chr16 | 79628537 | T | – | MAF | rs66467731 | 16q23.2 | ||
| Patient 1 | chr17 | 7578712 | TTTT | – | TP53 | 17p13.1 | |||
| Patient 1 | chr17 | 11924223 | GCG | – | MAP2K4 | Nonframeshift deletion | 17p12 | ||
| Patient 1 | chr17 | 43364293 | – | G | MAP3K14 | Unknown | rs56405343 | 17q21.31 | |
| Patient 1 | chrX | 47030561 | GGA | – | RBM10 | Nonframeshift deletion | Xp11.23 | ||
| Patient 1 | chrX | 53222043 | – | C | KDM5C | rs146836963 | Xp11.22 | ||
| Patient 1 | chrX | 66765158 | – | GCAGCA | AR | Nonframeshift insertion | Xq12 | ||
| Patient 2 | chr5 | 35861068 | T | C | IL7R | Nonsynonymous SNV | rs1494558 | Benign | 5p13.2 |
| Patient 2 | chr5 | 35871190 | G | A | IL7R | Nonsynonymous SNV | rs1494555 | Benign | 5p13.2 |
| Patient 2 | chr5 | 176520243 | G | A | FGFR4 | Nonsynonymous SNV | rs351855 | Benign | 5q35.2 |
| Patient 2 | chr12 | 121437382 | A | G | HNF1A | Nonsynonymous SNV | Benign | 12q24.31 |
Therefore, BCAR1 is a possible susceptibility gene for lung cancer. Although the biological function of BCAR1 in lung cancer remains unclear, we believe that BCAR1 plays an important role in the development and progression of lung cancer. It is worth noting that the 2 siblings developed 2 different histologic types of lung cancer, revealing that BCAR1 may use different mechanisms to induce different histologic types of lung cancer. To further support our hypothesis, investigation concerning the biological function and interaction protein network of BCAR1 in the occurrence and development of lung cancer is urgent and necessary. In addition, there were another 3 carriers identified who were in good health conditions, and a long-term follow-up is of great necessity.
In summary, our report revealed a possible susceptibility gene, BCAR1, and further investigation of biological function of BCAR1 in lung cancer is needed. Our study was the first to identify the frameshift germline mutation of BCAR1 by WES and Sanger sequencing, which is beneficial for the diagnosis and in the long term precision treatment of lung cancer.
Acknowledgments
The authors thank Shanghai Tongshu Biotechnology Co., Ltd. for technical support and the patients and their family for their interest and cooperation.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-7017/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-7017/coif). All authors report that this work had received technical support from Shanghai Tongshu Biotechnology Co., Ltd. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the ethical standards of the institutional and or national research committee(s) and with the Helsinki Declaration (as revised in 2013) and ethical standards of the Medical Ethics Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology and got its approval (ethical approval No. S412). Written informed consent was obtained from both patients for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer 2007;7:778-90. [Crossref] [PubMed]
- Gazdar A, Robinson L, Oliver D, et al. Hereditary lung cancer syndrome targets never smokers with germline EGFR gene T790M mutations. J Thorac Oncol 2014;9:456-63. [Crossref] [PubMed]
- Jonsson S, Thorsteinsdottir U, Gudbjartsson DF, et al. Familial risk of lung carcinoma in the Icelandic population. JAMA 2004;292:2977-83. [Crossref] [PubMed]
- Coté ML, Liu M, Bonassi S, et al. Increased risk of lung cancer in individuals with a family history of the disease: a pooled analysis from the International Lung Cancer Consortium. Eur J Cancer 2012;48:1957-68. [Crossref] [PubMed]
- Yu HA, Arcila ME, Harlan Fleischut M, et al. Germline EGFR T790M mutation found in multiple members of a familial cohort. J Thorac Oncol 2014;9:554-8. [Crossref] [PubMed]
- Yamamoto H, Higasa K, Sakaguchi M, et al. Novel germline mutation in the transmembrane domain of HER2 in familial lung adenocarcinomas. J Natl Cancer Inst 2014;106:djt338. [Crossref] [PubMed]
- Abdel-Rahman MH, Pilarski R, Cebulla CM, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet 2011;48:856-9. [Crossref] [PubMed]
- Wang Y, McKay JD, Rafnar T, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet 2014;46:736-41. [Crossref] [PubMed]
- Barrett A, Pellet-Many C, Zachary IC, et al. p130Cas: a key signalling node in health and disease. Cell Signal 2013;25:766-77. [Crossref] [PubMed]
- Defilippi P, Di Stefano P, Cabodi S. p130Cas: a versatile scaffold in signaling networks. Trends Cell Biol 2006;16:257-63. [Crossref] [PubMed]
- Camacho Leal Mdel P, Sciortino M, Tornillo G, et al. p130Cas/BCAR1 scaffold protein in tissue homeostasis and pathogenesis. Gene 2015;562:1-7. [Crossref] [PubMed]
- Imyanitov EN, Iyevleva AG, Levchenko EV. Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Crit Rev Oncol Hematol 2021;157:103194. [Crossref] [PubMed]
- Lou Y, Pecot CV, Tran HT, et al. Germline Mutation of T790M and Dual/Multiple EGFR Mutations in Patients With Lung Adenocarcinoma. Clin Lung Cancer 2016;17:e5-11. [Crossref] [PubMed]
- Lammers PE, Lovly CM, Horn L. A patient with metastatic lung adenocarcinoma harboring concurrent EGFR L858R, EGFR germline T790M, and PIK3CA mutations: the challenge of interpreting results of comprehensive mutational testing in lung cancer. J Natl Compr Canc Netw 2014;12:6-11; quiz 11. [Crossref] [PubMed]
- Wu Y, Sarkissyan M, Vadgama JV. Epigenetics in breast and prostate cancer. Methods Mol Biol 2015;1238:425-66. [Crossref] [PubMed]
- Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet 2012;13:679-92. [Crossref] [PubMed]
- Van Den Broeck A, Brambilla E, Moro-Sibilot D, et al. Loss of histone H4K20 trimethylation occurs in preneoplasia and influences prognosis of non-small cell lung cancer. Clin Cancer Res 2008;14:7237-45. [Crossref] [PubMed]
- Simó-Riudalbas L, Esteller M. Targeting the histone orthography of cancer: drugs for writers, erasers and readers. Br J Pharmacol 2015;172:2716-32. [Crossref] [PubMed]
- Hascher A, Haase AK, Hebestreit K, et al. DNA methyltransferase inhibition reverses epigenetically embedded phenotypes in lung cancer preferentially affecting polycomb target genes. Clin Cancer Res 2014;20:814-26. [Crossref] [PubMed]
- Mullapudi N, Ye B, Suzuki M, et al. Genome Wide Methylome Alterations in Lung Cancer. PLoS One 2015;10:e0143826. [Crossref] [PubMed]
- Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A 2007;104:15805-10. [Crossref] [PubMed]
- Frebourg T. The challenge for the next generation of medical geneticists. Hum Mutat 2014;35:909-11. [Crossref] [PubMed]
- Xue Y, Ankala A, Wilcox WR, et al. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet Med 2015;17:444-51. [Crossref] [PubMed]
- Boycott KM, Vanstone MR, Bulman DE, et al. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet 2013;14:681-91. [Crossref] [PubMed]
- Zhang X. Exome sequencing greatly expedites the progressive research of Mendelian diseases. Front Med 2014;8:42-57. [Crossref] [PubMed]
- Krauthammer M, Kong Y, Ha BH, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet 2012;44:1006-14. [Crossref] [PubMed]
- Liu P, Morrison C, Wang L, et al. Identification of somatic mutations in non-small cell lung carcinomas using whole-exome sequencing. Carcinogenesis 2012;33:1270-6. [Crossref] [PubMed]
- Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012;482:226-31. [Crossref] [PubMed]
- Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011;478:64-9. [Crossref] [PubMed]
- Yan XJ, Xu J, Gu ZH, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet 2011;43:309-15. [Crossref] [PubMed]
- Chang H, Jackson DG, Kayne PS, et al. Exome sequencing reveals comprehensive genomic alterations across eight cancer cell lines. PLoS One 2011;6:e21097. [Crossref] [PubMed]
- Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 2011;469:539-42. [Crossref] [PubMed]
- Bignell GR, Greenman CD, Davies H, et al. Signatures of mutation and selection in the cancer genome. Nature 2010;463:893-8. [Crossref] [PubMed]
- Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 2010;463:360-3. [Crossref] [PubMed]
- Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 2010;42:181-5. [Crossref] [PubMed]
- Tagliabracci VS, Engel JL, Wen J, et al. Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science 2012;336:1150-3. [Crossref] [PubMed]
- Deng B, Huang W, Tan QY, et al. Breast cancer anti-estrogen resistance protein 1 (BCAR1/p130cas) in pulmonary disease tissue and serum. Mol Diagn Ther 2011;15:31-40. [Crossref] [PubMed]
- Wolpin BM, Rizzato C, Kraft P, et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet 2014;46:994-1000. [Crossref] [PubMed]
- Li Y, Wang X, Vural S, et al. Exome analysis reveals differentially mutated gene signatures of stage, grade and subtype in breast cancers. PLoS One 2015;10:e0119383. [Crossref] [PubMed]
- Meijer D, van Agthoven T, Bosma PT, et al. Functional screen for genes responsible for tamoxifen resistance in human breast cancer cells. Mol Cancer Res 2006;4:379-86. [Crossref] [PubMed]
- Brinkman A, van der Flier S, Kok EM, et al. BCAR1, a human homologue of the adapter protein p130Cas, and antiestrogen resistance in breast cancer cells. J Natl Cancer Inst 2000;92:112-20. [Crossref] [PubMed]
- Cabodi S, Tinnirello A, Di Stefano P, et al. p130Cas as a new regulator of mammary epithelial cell proliferation, survival, and HER2-neu oncogene-dependent breast tumorigenesis. Cancer Res 2006;66:4672-80. [Crossref] [PubMed]
- Huang W, Deng B, Wang RW, et al. BCAR1 protein plays important roles in carcinogenesis and predicts poor prognosis in non-small-cell lung cancer. PLoS One 2012;7:e36124. [Crossref] [PubMed]
- Nick AM, Stone RL, Armaiz-Pena G, et al. Silencing of p130cas in ovarian carcinoma: a novel mechanism for tumor cell death. J Natl Cancer Inst 2011;103:1596-612. [Crossref] [PubMed]
- Jiang S, Mao C, Jiang B, et al. High Expression of BCAR1 by Circulating Tumor Cells and Tumor Tissues Is Predictive of a Poor Prognosis of Early-Stage Lung Adenocarcinoma Potentially Due to Regulation of Epithelial-Mesenchymal Transition. Technol Cancer Res Treat 2020;19:1533033820983086. [Crossref] [PubMed]
- Chodniewicz D, Klemke RL. Regulation of integrin-mediated cellular responses through assembly of a CAS/Crk scaffold. Biochim Biophys Acta 2004;1692:63-76. [Crossref] [PubMed]
- Brábek J, Constancio SS, Siesser PF, et al. Crk-associated substrate tyrosine phosphorylation sites are critical for invasion and metastasis of SRC-transformed cells. Mol Cancer Res 2005;3:307-15. [Crossref] [PubMed]
- Sanders MA, Basson MD. p130cas but not paxillin is essential for Caco-2 intestinal epithelial cell spreading and migration on collagen IV. J Biol Chem 2005;280:23516-22. [Crossref] [PubMed]
- Wei L, Yang Y, Zhang X, et al. Cleavage of p130Cas in anoikis. J Cell Biochem 2004;91:325-35. [Crossref] [PubMed]
- Kim W, Kook S, Kim DJ, et al. The 31-kDa caspase-generated cleavage product of p130cas functions as a transcriptional repressor of E2A in apoptotic cells. J Biol Chem 2004;279:8333-42. [Crossref] [PubMed]
- Ambrogio C, Voena C, Manazza AD, et al. p130Cas mediates the transforming properties of the anaplastic lymphoma kinase. Blood 2005;106:3907-16. [Crossref] [PubMed]
- Cabodi S, Moro L, Baj G, et al. p130Cas interacts with estrogen receptor alpha and modulates non-genomic estrogen signaling in breast cancer cells. J Cell Sci 2004;117:1603-11. [Crossref] [PubMed]
- Makkinje A, Vanden Borre P, Near RI, et al. Breast cancer anti-estrogen resistance 3 (BCAR3) protein augments binding of the c-Src SH3 domain to Crk-associated substrate (p130cas). J Biol Chem 2012;287:27703-14. [Crossref] [PubMed]
- Zhang XA, He B, Zhou B, et al. Requirement of the p130CAS-Crk coupling for metastasis suppressor KAI1/CD82-mediated inhibition of cell migration. J Biol Chem 2003;278:27319-28. [Crossref] [PubMed]
- Heumann A, Heinemann N, Hube-Magg C, et al. High BCAR1 expression is associated with early PSA recurrence in ERG negative prostate cancer. BMC Cancer 2018;18:37. [Crossref] [PubMed]
- Deng B, Sun Z, Jason W, et al. Increased BCAR1 predicts poor outcomes of non-small cell lung cancer in multiple-center patients. Ann Surg Oncol 2013;20:S701-8. [Crossref] [PubMed]
- Mao CG, Jiang SS, Shen C, et al. BCAR1 promotes proliferation and cell growth in lung adenocarcinoma via upregulation of POLR2A. Thorac Cancer 2020;11:3326-36. [Crossref] [PubMed]


