
A novel variant of SPAST in a pedigree with pure hereditary spastic paraplegia in Yunnan Province
Introduction
Hereditary spastic paraplegia (HSP), also known as Strümpell-Lorrain Syndrome, is a rare group of genetically heterogeneous neurodegenerative disorders with a prevalence of 4.3–9.8/100,000, depending on the ethnic population (1). The clinical manifestations are mainly progressive spastic gait, increased muscle tension at rest of both lower limbs, hyperreflexia of the tendon, clonic spasms of the ankle, positive pathological signs, and myasthenia gravis, possibly accompanied by abnormal bladder and rectal function, ankle joint vibration loss, clawfoot, and scoliosis (2,3). HSP can start before 35 years of age (early onset) or after 35 years of age (classical) and can classified as pure (uncomplicated) and complicated (complex) based on signs and symptoms, according to Harding’s criteria outlined in 1981 and 1983 (2,4-6). The pure form is confined to the progressive spastic paresis with clinical manifestations restricted to corticospinal system degeneration, including lower extremity weakness and spasticity, corticospinal tract signs, disturbance in vibration sense and proprioception, and a variable hypertonic urinary disturbance (5-8). Complicated HSP is involved with additional neurological signs (8), including mental retardation, cerebellar ataxia, intellectual deficiency, epilepsy, peripheral neuropathy, optic atrophy, retinitis pigmentosa, nervous system dysfunction, and other extra-pyramidal features, such as neurological deafness, white matter lesion (WML) and thin corpus callosum (TCC) (9).
HSP can have autosomal dominant (AD), autosomal recessive (AR), or X-linked inheritance, and at least 80 loci and 79 genes have been identified as HSP subtypes (10-12). With respect to the genetic basis of HSP, the majority of HSP cases (approximately 80%) are characterized as AD, and more than 20 AD-HSP spastic paraplegia (SPG) subtypes have been identified (including SPG3A, SPG4, SPG6, SPG8, SPG9, SPG10, SPG37, and SPG73,etc.). Among them, hereditary spastic paraplegia type 4 (SPG4) is the most frequent subtype of AD-HSP (SPG4, MIM: 182601) and comprises 45% of HSP. As the most frequent causal gene of SPG4, the SPG4/SPAST (protein: Spastin) gene accounts for approximately 40% of AD pedigrees and 20% of sporadic cases, respectively (13). In addition, more than 45 autosomal recessive (AR)-HSP subtypes have been identified (including SPG5, SPG7, SPG11, SPG14, and SPG15, etc.) (5,14), and variants in SPG5, SPG11, and SPG15 have been described as the most frequent causes for AR-HSP (15) .
SPG4 as the commonest AD HSP form in many population, although de novo variations do appear, most SPAST-HSP cases are hereditary (16-18). It has been reported that the average onset of AD in SPG4/SPAST patients was 24.79 (95% CI: 21.00–28.58) (19) or mostly in their fourth decade (20). Recently, a meta-analysis on 13,570 HSP patients found that a pooled frequency of variations in SPAST was 25% (95% CI: 21.00–30.00%), and no significant difference frequencies among different populations (Q=3.47, P=1.00); the frequency of variations in the Asian population (32.62%, 95% CI: 23.93–42.70%) was higher than that of Caucasians (23.07%, 95% CI: 18.73–28.06%) and Americans (24.83%, 95% CI: 15.59–37.05%) (19).
In this study, we presented a four-generation pedigree of HSP patients and performed whole-exome sequencing (WES) to screen the pathological candidate gene(s) and variant(s). SPAST with a novel nonsense variation (c.1510 C>T, p.Gln504X, Ref Seq. NM_199436) showing an AD inheritance pattern was identified in the pedigree, and in function prediction it was regarded as a causally associated gene through the combination of clinical features. These findings fit in well with the known spectrum of SPG4 and were not reported in the literature.
We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6698/rc).
Methods
Subjects
The present study involved a large pedigree cohort of 11 HSP patients in Yunnan Province (Figure 1). Biochemical tests, electromyography (EMG), and brain and thoracic spinal cord magnetic resonance imaging (MRI) were conducted for seven individuals (Cases 1, 2, 3, 6, 8, 10, and 14). The detailed clinical analysis of pure HSP in this pedigree is summarized in Table 1. In brief, the 7 individuals were diagnosed as HSP patients on the basis of Harding’s criteria (2,4): all of them harbored symptoms which manifested as weakness of the lower limbs, hyperactivity of the tendon reflex, positive pathological reflex, and ataxia. There was no presentation of symptoms of abnormal higher-level mental activity, epileptic scabs, skin lesions, limb muscle atrophy and paresthesia, diminution of vision, and cataracts. In addition to the obvious structural and signal abnormalities observed via MRI, the symptoms of all patients were consistent with the manifestation of pure HSP. This pedigree cohort had no close-relative marriage history. Furthermore, there was no history of birth injury.
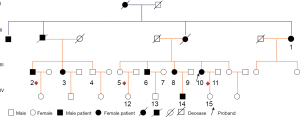
Table 1
| Symptoms and genotypes | Cases of HSP patients | Cases of controls | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1* | 2 | 3 | 6 | 8 | 10 (P) | 14 | 4 | 5 | 7 | 9 | 11 | 12 | 13 | 15 | ||
| Onset age | 67 | 30 | 32 | 26 | 33 | 10 | 20 | − | − | − | − | − | − | − | − | |
| Gender (female/male) | F | M | F | M | F | F | M | − | − | − | − | − | − | − | − | |
| Spastic gait | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | |
| Dysuria | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Postural tremor | − | − | − | + | + | − | − | − | − | − | − | − | − | − | − | |
| Upper limbs weakness | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Upper limbs hypermyotonia | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | |
| Upper limbs hyperreflexia | − | − | − | Active | Active | Active | Active | − | − | − | − | − | − | − | − | |
| Lower limb weakness | + | + | + | + | + | + | − | − | − | − | − | − | − | − | − | |
| Lower limbs hypermyotonia | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | |
| Lower limbs hyperreflexia | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | |
| Babinski sign | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − | |
| Cognitive disorder | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Paresthesia | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Scoliosis | + | + | − | − | − | + | − | − | − | − | − | − | − | − | − | |
| Claw foot | + | − | − | − | − | + | − | − | − | − | − | − | − | − | − | |
| Lower limbs motor function grading | + | + | − | − | − | − | − | − | − | − | ||||||
| Grade I (normal/abnormal gait) | + | − | − | − | − | − | − | − | − | |||||||
| Grade II (ambulation without jump) | + | + | + | − | − | − | − | − | − | − | − | |||||
| Grade III (dependent ambulation) | + | − | − | − | − | − | − | − | − | |||||||
| Grade IV (wheel-dependent/bedridden) | ||||||||||||||||
| Genotypes | ||||||||||||||||
| SPAST (novel, NM 199436, exon 13, c.1510 C>T,p.Q504X) | C/T | C/T | C/T | C/T | C/T | C/T | C/T | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/C | |
| CPT1C (rs150853576, NM_001199752, exon14, c.1541G>T, p.R514L) | G/T | G/T | G/T | G/G | G/T | G/T | G/T | G/G | G/G | G/G | G/G | G/G | G/G | G/G | G/G | |
| DNAJC16 (novel, NM 015291.4, exon5, c.718 C>T, p.Q240X) | C/C | C/T | C/T | C/T | C/T | C/T | C/T | C/C | C/C | C/C | C/C | C/C | C/C | C/C | C/T | |
*, presented almost no symptoms of HSP at the time of first medical assessment (2012), gradually progressed into a typical HSP patient during follow-up in 2018. “P” means proband. “+” means positive; ”−” means negative in HSP patients or did not check in the controls. HSP, hereditary spastic paraplegia; F, female; M, male; A, active.
The recruitment and detection were implemented by the Neurology Department and the Clinical Basic Medical Institute of the First People’s Hospital of Yunnan Province. Blood samples of the 7 patients and another 8 phenotype normal controls (Cases 4, 5, 7, 9, 11, 12, 13, and 15) were collected for DNA extraction (Cat# DP1802; Tiangen Biotech Co., Ltd, Beijing, China) and laboratory examination. In addition, blood samples from another 300 phenotype normal controls were collected to verify the candidate variants. Oral informed consent was taken from all the patients or their families. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was conducted with approval of the Ethics Committee of the First People’s Hospital of Yunnan Province (YYLH097).
WES sequencing
WES was performed on two affected (Cases 2, 10) and one control (Case 5) individuals. Accordingly, the DNA was fragmented to an average size of 180–280 base pairs (bp) and used to create a DNA library, following the established Illumina paired-end protocols. Then, the SureSelect Human All Exon v 6.0 Kit (Agilent Technologies, Santa Clara, CA, USA) was used for exome capture, according to the manufacturer’s instructions. WES was executed on the Illumina HiSeq X platform (Illumina Inc., San Diego, CA, USA) based on the Human Sequencing Library and HiSeq XTen whole genome sequencing software (Novogene Bioinformatics Technology Co., Ltd., Beijing, China). The average raw data flux per sample was not less than 90 G with a raw depth of 30X, and 150 bp paired-end reads were generated.
Variant filtration process
After the sequencing, base-call file conversion and demultiplexing were performed using the bcl2fastq software (Illumina, San Diego, CA, USA). The raw data was analyzed by an in-house quality control software to remove adaptor reads, reads containing N sequences, and low-quality reads, and to align these to the reference human genome (GRCh37/hg19). Then, the results that referred to a single nucleotide variant (SNV) from these samples were obtained using SAMtools software v. 1.0, and these samples were subsequently combined and merged for annotation, according to their own virtual contact file (VCF). Then, the filtering of candidate genes and variants was performed as follows: (I) variants with a minor allele frequency (MAF) value <0.01 in 1000 genomic data (1000g_all), ExAC (Exome Aggregation Consortium) data (ExAC _ALL) and the in-house Novo-Zhonghua exome database from Novogene (NovoDb_WGS_SNP) were obtained; (II) SNVs occurring in exons or splice sites (splicing junction 10 bp) were further analyzed; (III) synonymous SNVs predicted by the Database of Splicing Consensus Single-Nucleotide Variants (dbscSNV) were discarded, and small fragment non-frameshift (<10 bp) indels in the repeat regions defined by RepeatMasker software (version. 4.1.0) were also discarded; (IV) the variations were screened according to the scores using the Sorting Tolerant from Intolerant (SIFT) (version 2.6), Polyphen (http://genetics.bwh.harvard.edu/pph2/), MutationTaster (http://www.mutationtaster.org/), and computer-aided design and drafting (CADD) software programs (v1.5). Potentially deleterious variations were retained when the score from more than half of these four software programs supported their potential harmfulness. Sites (>2 bp) that did not affect the alternative splicing were removed; (V) candidate genes and variants were screened on the basis of the dominant inheritance pattern.
Verification by Sanger sequencing
Sanger sequencing was carried out to validate the candidate variants on an ABI 3500xL Dx Genetic Analyzer (Applied Biosystems, Foster City, USA). The primers of the candidate genes were available upon request.
Results
Clinical progression of pedigree members
Of the 15 enrolled members, Case 1, who had presented with almost no symptoms of HSP at the time of their first medical assessment, gradually progressed into a typical HSP patient during follow up (Figure 1). The rest of the members showed no other obvious symptoms of progression for the time being, except the affected individuals (Cases 2, 3, 6, 8, 10, and 14) who showed signs of gradually severe spastic paraplegia of both lower limbs, hyperactivity of the tendon reflex, and progressive instability of gait, consistent with HSP patients (Table 1).
Considering that the proband Case 10 showed HSP symptoms at the earliest age of ten years old, Case 14 experienced early-age onset of HSP at twenty years old, and Case 1 involved a recently confirmed female case of HSP, it seems that the onset age (31.1±17.7) of this HSP pedigree ranges across a long time span due to the genetic crosstalk.
WES results and sequencing analysis
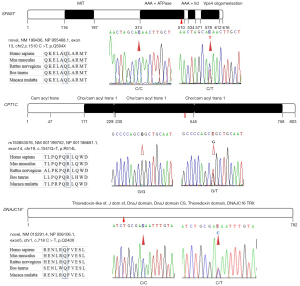
After the raw reads were filtered, 620,043,626 (Case 2), 612,419,612 (Case 5), and 613,997,406 (Case 10) clean reads were obtained with an average sequencing depth of 30.89×. A total of 609,793,398 (Case 2), 608,654,099 (Case 5), and 612,901,275 (Case 10) reads were mapped with GRCh37/hg19 (99.39–99.82%), which indicated that the assembled unigenes could be used for subsequent analysis. The numbers of SNVs within coding range were 3,572,793 (Case 2), 3,606,691 (Case 5), and 3,595,000 (Case 10). After filtering, 2 SPG-related genes (SPAST and CPT1C) and DNAJC16 with or without novel variations were filtered as candidate genes and verified by Sanger sequencing as belonging to this pedigree (Figure 2, Table 1). Only SPAST with a novel stop-gain variant (c.1510 C>T, p.Gln504X, RefSeq.NM_199436) fitted the AD pattern; none of this variant was found in the control groups, except for the seven HSP patients (Table 1). The accession numbers of the novel variants in the ClinVar database were SCV001573094 and SCV001573804, respectively.
Discussion
The SPAST gene, also known as the SPG4 gene, has been mapped to chromosome 2p21-p22, and encodes spastin, an amino acid protein belonging to the broad AAA (triple A; ATPase associated with various cellular activities) family (16). With two initiation codons, SPG4 mRNA synthesizes two spastin isoforms, a 616-amino acid (68 kDa) isoform called M1 and a 530-amino acid (60 kDa) isoform called M87. These isoforms have a different subcellular localization, with the short one being present in relevant amounts both in the cytoplasm and the nucleus, and the long one being excluded from the nucleus by active export (21). Spastin display microtubule- severing ATPase activity and uses energy from ATP hydrolysis to sever and disassemble microtubules; disease variations abolish or partially interfere with these activities (21,22). According to the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/gene.php?gene=SPAST), over 600 SPAST variations have been identified, including missense, nonsense, deletions, insertions, and splice-site variations (23-27). In this pedigree, the c.1510 C>T (p.Gln504X, RefSeq.NM_199436) variant in exon 13, which leads to a predicted premature termination and an 18% deletion of normal-size spastin, was confirmed to be evident in all HSP patients and transmitted among the pedigree via a definite AD hereditary manner. This stop-gain variant was not previously reported and was not seen in existing SNP databases, including gnomAD and ExAC. Spastin was mainly composed of TM (aa57-79), MIT (aa116-194), MTB (aa 270-328), AAA+ ATPase (aa374-510), AAA+ lid (aa534-571), and Vps4 oligomerisation, C-terminal (Vps4_C) (aa578-612) domains by in silico using the prediction programs InterPro (http://www.ebi.ac.uk/interpro/result/InterProScan/iprscan5-R20211227-062728-0240-35083180-p2m/). Previous research indicated that the TM/MIT region is involved in membrane targeting and the MIT/AAA region has activity on the microtubule cytoskeleton. Currently, more than 15 variants clustering the AAA+ ATPase domain were reported by HSP patients (17,25). The nonsense variation identified in this pedigree clustering in the AAA+ ATPase domain, and results in the deletion of the predicted AAA+ lid domain and the Vps4_C domain. In addition, Mutation Taster software in silico predicted that this variant was a “disease causing” variation, thus suggesting the pathogenicity of c.1510 C>T (p.Gln504X, RefSeq.NM_199436) in this pedigree. To date, loss-of-function (haploinsufficiency) and gain-of function have been the most prevalent explanation for SPAST-based HSP (28-30). In fact, several truncated variants of spastin showed impaired microtubule-severing activity and neurotoxicity in vitro. Published data showed that the truncated spastin exerts an isoform-specific effect on microtubule dynamics (31,32). Recent experiments showed that nonsense variants of SPAST, such as Asn184X, Ser245X and Met329X, produce two truncated variants (mutant M1 and M87 isoforms) that accumulated to a higher level than their wild-type counterparts. The mutate M1 isoform heavily decorated the microtubules and rendered them resistant to depolymerization. In contrast, the mutate M87 isoform could not decorate microtubules and was not able to promote microtubule disassembly (31,32). However, the reported artificial truncated proteins using in in vitro experiments completely lack the AAA domain and are shorter than the variant found in our patients, thus, further studies on the exact mechanism of its haploinsufficiency or gain-of function are certainly necessary.
Furthermore, a reported SNV of SPG73 was also tested in this pedigree. The SPG73 was previously identified in a family with variants in the neuronal isoform of the carnitine palmitoyl-transferase 1C (CPT1C) gene (33). The study identified the nucleotide substitution c.109C>T in exon 3 of CPT1C, and expanded the genetics of the pure form of AD-HSP. Consistently, our study found the rs150853576 variant covered almost all HSP patients in an AD hereditary manner, except for Case 6, which had no publications for rs150853576, except for the following research: https://www.ncbi.nlm.nih.gov/snp/rs150853576?vertical_tab=true#publications (date of consultation Dec. 2021). However, the heterozygous and homozygous variant of this SNP were also found in 300 controls (3/300 vs. 1/300), respectively. In fact, a recent study by Hong et al. found a novel heterozygous variant (c.226C>T) resulting in a premature stop (p.Q76X) in exon 3 of the CPT1C, with benign clinical course (34). Therefore, whether the rs150853576 variant might be one of the “minor offenders” to contribute to cumulative risk and the occurrence of pure HSP in this pedigree, needs further functional verification.
Interestingly, a novel c.718 C>T (p.Q240X, Ref Seq NM_015291) variant in exon 5 of DNAJC16 resulted in a predicted protein that was not rapidly degraded, was potentially truncated by 69% its normal size, and was identified in almost all HSP individuals, except for Case 1 in this pedigree. DNAJC16 was located on chromosome 1p36.21 and encoded ERdj8, a 782-amino-acid protein. Recent reports revealed that ERdj8 localizes to a meshwork-like ER subdomain. Along with phosphatidylinositol synthase (PIS) and the autophagy-related (Atg) proteins, it is associated with the regulation of the size of autophagosomes. Its over expression increased the diameter of the autophagosomes in non-selective autophagy, and its ablation resulted in a defect in engulfing larger targets (35,36). Thus, considering that only Case 1 as an HSP patient did not bear this nonsensical variant of DNAJC16, and her clinical features were newly progressed, it is worth fast-tracking whether this predicted pathological variant has a synergistic effect on the clinical progress of this HSP pedigree.
As for the pedigree, the 7 investigated HSP individuals carried most of the pathological gene variants, including Case 1 (2/3), 2 (3/3), 3 (3/3), 6 (2/3), 8 (3/3), 10 (3/3), and 14 (3/3). Of significance is that Case 2 and Case 10 were selected for WES sequencing, with the result that both cases carried all the pathological variants so that the sequencing verification was credible. It was noteworthy that Case 8 also carried all the three variants, which was probably of the most relevance with her “Active” hyperreflexia in the upper limbs and dependent ambulation, diagnosed as Grade III (Table 1). Moreover, as Case 2 presented the most severe motor dysfunction as Grade IV (Table 1), and Case 3 as twin sister of Case 2 presented mild motor dysfunction as Grade I, indicating the clinical symptoms and time of onset are highly heterogeneous.
Conclusions
On the whole, most of the HSP patients who presented with possible pathological variants confirmed that the AD hereditary manner prevailed in this pure HSP pedigree. The SPAST (c.1510 C>T, p.Gln504X, RefSeq.NM_199436) as a genotype-phenotype cosegregated variant might be the “principal offender” of this pedigree. Considering the age of onset and clinical features, the other two variants might present cumulative risks to the occurrence and progression of HSP, to some genetic extent. However, further abundant applied studies should be carried out to obtain a more profound and sound understanding of the mechanisms used among these AD-HSP-related gene variants. Once further confirmation is achieved, these pathological variants or genes may potentially be applied as biomarkers in the diagnosis and provision of therapy during clinical treatment for pure HSP patients.
Acknowledgments
We thank the patients and their families for giving their consent to participate and donate samples. We thank Qian Zhao, Jiangyan Zhai, Ting Chen, and Peng Zeng for their expert technical assistance.
Funding: This work was supported by the Yunnan Provincial Science and Technology Department (2014FB086, 202001AY070001-109) to KWZ; in part by Yunnan Provincial Education Department (YJS-SJ-01) to KWZ; in part by the National Natural Science Foundation of China (81360102, 81160352) to TS; and in part by the Yunnan Health Commission (L2019003) to TS. The funding bodies provided us with financial support for data analysis and interpretation, and administrative work.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6698/rc
Data Sharing Statement: Available https://atm.amegroups.com/article/view/10.21037/atm-21-6698/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6698/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was conducted with approval of the Ethics Committee of the First People’s Hospital of Yunnan Province (YYLH097). Oral informed consent was taken from all the patients or their families.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lu C, Li LX, Dong HL, et al. Targeted next-generation sequencing improves diagnosis of hereditary spastic paraplegia in Chinese patients. J Mol Med (Berl) 2018;96:701-12. [Crossref] [PubMed]
- Harding AE. Hereditary "pure" spastic paraplegia: a clinical and genetic study of 22 families. J Neurol Neurosurg Psychiatry 1981;44:871-83. [Crossref] [PubMed]
- Morais S, Raymond L, Mairey M, et al. Massive sequencing of 70 genes reveals a myriad of missing genes or mechanisms to be uncovered in hereditary spastic paraplegias. Eur J Hum Genet 2017;25:1217-28. [Crossref] [PubMed]
- Harding AE. Classification of the hereditary ataxias and paraplegias. Lancet 1983;1:1151-5. [Crossref] [PubMed]
- Murala S, Nagarajan E, Bollu PC. Hereditary spastic paraplegia. Neurol Sci 2021;42:883-94. [Crossref] [PubMed]
- de Souza PVS, de Rezende Pinto WBV, de Rezende Batistella GN, et al. Hereditary Spastic Paraplegia: Clinical and Genetic Hallmarks. Cerebellum 2017;16:525-51. [Crossref] [PubMed]
- Parodi L, Fenu S, Stevanin G, et al. Hereditary spastic paraplegia: More than an upper motor neuron disease. Rev Neurol (Paris) 2017;173:352-60. [Crossref] [PubMed]
- Polo JM, Calleja J, Combarros O, et al. Hereditary "pure" spastic paraplegia: a study of nine families. J Neurol Neurosurg Psychiatry 1993;56:175-81. [Crossref] [PubMed]
- Tesson C, Koht J, Stevanin G. Delving into the complexity of hereditary spastic paraplegias: how unexpected phenotypes and inheritance modes are revolutionizing their nosology. Hum Genet 2015;134:511-38. [Crossref] [PubMed]
- Walusinski O. A historical approach to hereditary spastic paraplegia. Rev Neurol (Paris) 2020;176:225-34. [Crossref] [PubMed]
- Faber I, Pereira ER, Martinez ARM, et al. Hereditary spastic paraplegia from 1880 to 2017: an historical review. Arq Neuropsiquiatr 2017;75:813-8. [Crossref] [PubMed]
- Souza PVS, Bortholin T, Dias RB, et al. New genetic causes for complex hereditary spastic paraplegia. J Neurol Sci 2017;379:283-92. [Crossref] [PubMed]
- Zhu Z, Zhang C, Zhao G, et al. Novel mutations in the SPAST gene cause hereditary spastic paraplegia. Parkinsonism Relat Disord 2019;69:125-33. [Crossref] [PubMed]
- Burguez D, Polese-Bonatto M, Scudeiro LAJ, et al. Clinical and molecular characterization of hereditary spastic paraplegias: A next-generation sequencing panel approach. J Neurol Sci 2017;383:18-25. [Crossref] [PubMed]
- Schüle R, Wiethoff S, Martus P, et al. Hereditary spastic paraplegia: Clinicogenetic lessons from 608 patients. Ann Neurol 2016;79:646-58. [Crossref] [PubMed]
- Feng Y, Ke X, Zhai M, et al. Novel deletion of SPAST in a Chinese family with hereditary spastic paraplegia. Singapore Med J 2013;54:251-4. [Crossref] [PubMed]
- de Bot ST, van den Elzen RT, Mensenkamp AR, et al. Hereditary spastic paraplegia due to SPAST mutations in 151 Dutch patients: new clinical aspects and 27 novel mutations. J Neurol Neurosurg Psychiatry 2010;81:1073-8. [Crossref] [PubMed]
- Nan H, Okamoto K, Gao L, et al. A Japanese SPG4 Patient with a Confirmed De Novo Mutation of the SPAST Gene. Intern Med 2020;59:2311-5. [Crossref] [PubMed]
- Erfanian Omidvar M, Torkamandi S, Rezaei S, et al. Genotype-phenotype associations in hereditary spastic paraplegia: a systematic review and meta-analysis on 13,570 patients. J Neurol 2021;268:2065-82. [Crossref] [PubMed]
- Lo Giudice T, Lombardi F, Santorelli FM, et al. Hereditary spastic paraplegia: clinical-genetic characteristics and evolving molecular mechanisms. Exp Neurol 2014;261:518-39. [Crossref] [PubMed]
- Claudiani P, Riano E, Errico A, et al. Spastin subcellular localization is regulated through usage of different translation start sites and active export from the nucleus. Exp Cell Res 2005;309:358-69. [Crossref] [PubMed]
- Roll-Mecak A, Vale RD. The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr Biol 2005;15:650-5. [Crossref] [PubMed]
- Morikawa T, Miura S, Tateishi T, et al. A Japanese hereditary spastic paraplegia family with a rare nonsynonymous variant in the SPAST gene. Hum Genome Var 2021;8:21. [Crossref] [PubMed]
- Yu W, Jin H, Deng J, et al. A novel SPAST gene mutation identified in a Chinese family with hereditary spastic paraplegia. BMC Med Genet 2020;21:123. [Crossref] [PubMed]
- Robbins NM, Ozmore JR, Winder TL, et al. A Novel SPAST/SPG4 Splice-Site Variant in a Family with Dominant Hereditary Spastic Paraplegia. Case Rep Neurol Med 2020;2020:7219514. [Crossref] [PubMed]
- She H, Zheng X, Xiao Y, et al. Novel SPG4 Mutation in a Patient with Sporadic Hereditary Spastic Paraplegia and Elevated Cerebrospinal Fluid Protein. J Clin Neurol 2021;17:152-3. [Crossref] [PubMed]
- Angelini C, Goizet C, Said SA, et al. Evidence of mosaicism in SPAST variant carriers in four French families. Eur J Hum Genet 2021;29:1158-63. [Crossref] [PubMed]
- Solowska JM, D'Rozario M, Jean DC, et al. Pathogenic mutation of spastin has gain-of-function effects on microtubule dynamics. J Neurosci 2014;34:1856-67. [Crossref] [PubMed]
- Solowska JM, Garbern JY, Baas PW. Evaluation of loss of function as an explanation for SPG4-based hereditary spastic paraplegia. Hum Mol Genet 2010;19:2767-79. [Crossref] [PubMed]
- Mohan N, Qiang L, Morfini G, et al. Therapeutic Strategies for Mutant SPAST-Based Hereditary Spastic Paraplegia. Brain Sci 2021;11:1081. [Crossref] [PubMed]
- Chen R, Du S, Yao Y, et al. A Novel SPAST Mutation Results in Spastin Accumulation and Defects in Microtubule Dynamics. Mov Disord 2021; [Epub ahead of print]. [Crossref] [PubMed]
- Solowska JM, Rao AN, Baas PW. Truncating mutations of SPAST associated with hereditary spastic paraplegia indicate greater accumulation and toxicity of the M1 isoform of spastin. Mol Biol Cell 2017;28:1728-37. [Crossref] [PubMed]
- Rinaldi C, Schmidt T, Situ AJ, et al. Mutation in CPT1C Associated With Pure Autosomal Dominant Spastic Paraplegia. JAMA Neurol 2015;72:561-70. [Crossref] [PubMed]
- Hong D, Cong L, Zhong S, et al. A novel CPT1C variant causes pure hereditary spastic paraplegia with benign clinical course. Ann Clin Transl Neurol 2019;6:610-4. [Crossref] [PubMed]
- Yamamoto YH, Kasai A, Omori H, et al. ERdj8 governs the size of autophagosomes during the formation process. J Cell Biol 2020;219:e201903127. [Crossref] [PubMed]
- Yamamoto YH, Noda T. Autophagosome formation in relation to the endoplasmic reticulum. J Biomed Sci 2020;27:97. [Crossref] [PubMed]
(English Language Editor: K. Gilbert)


