
Homology modeling and molecular docking simulation of martentoxin as a specific inhibitor of the BK channel
Introduction
Large-conductance calcium-activated potassium channels (BK channels) are gated by both voltage and calcium ions (1,2), and are widely distributed in excitable and nonexcitable cells. Each BK channel has 4 identical alpha subunits encoded by the Slo1 gene (3,4) and an ionic pore with an activation gate, a voltage sensor and a large C-terminal domain (5). In some subtypes of BK channels, there are auxiliary subunits (β and γ subunits) involved in maintaining the physiological functions of channels. Different combinations of α, β and γ subunits form BK subtypes with district physiological functions.
BK channels are key participants in a variety of physiological functions, such as regulating cell excitability and controlling cell proliferation, migration, and osmotic pressure. Dysfunction of the BK channel often induces a series of diseases. In vascular smooth muscle cells, reducing the Ca2+ sensitivity of the BK channel was shown to induce high blood pressure (6,7). In myocardial cells, the BK channel, which is located in the mitochondrial membrane, can prevent the damage caused by ischemia and hypoxia in the myocardium (8). In neuronal cells, the absence or downregulation of BK channels can lead to a series of diseases, such epilepsy, dyskinesia-associated diseases, and deafness (9-12). The expression of the BK channel is closely related to the degree of malignancy of glial cells in glioma. Exploring the mechanism underlying the interaction between the ligand and the BK channel with a specific ligand could provide insights into the function of BK channels.
Analyzing the structure of channels is of great significance to reveal the function of channels. It is reported that the structure of several potassium channel families has been elucidated, including the Kv1.2 channel and K+ Conduction and Selectivity Architecture (KcsA) (13,14). However, the structure of BK channel has not been fully analyzed.
It is an efficient method to study the structural-function by using the interaction between specific drug tools and specific structures of membrane channels. Currently, specific agonists and inhibitors are the most commonly used tool drugs in the functional study of BK channel subtypes. Frequently-used agonists could be mainly divided into three categories: (I) BK channel agonists (DHS-1, DiBAC4) that depend on the presence of β subunits, DHS-1 can activate BK channels composed of α and β1 subunits (15,16); (II) BK channel agonists (Maxikdiol and Pimaric acid) dependent on deletion of β subunits, Maxikido1 can only activate the BK channel expressed by α subunit alone (17); (III) BK channel agonists unrelated to the presence of β subunits [4-Chloro-7-trifluoromethyl-10H-benzo[4,5] furo [3,2-b] indole-1-carboxylic acid (TBIC)] (18). BK channel inhibitors mainly include the polypeptide toxins [charybdotoxin (ChTX), Iberiotoxin (IbTx) and Martentoxin] and non-peptide toxins (Paxilline, Penitrem A, Paspalinine and Tetrandrine) (19,20). Non-peptide toxins have low specificity for BK channels, whereas polypeptide toxoid can specifically inhibit BK channel activity at low concentration. Therefore, polypeptide toxins are more commonly used to study the structure-functional properties of BK channel subtypes. For instance, ChTX is sensitive to BK channel formed by α subunit expression alone. The co-expression of β1 and α subunits of BK channel increased the sensitivity of ChTX to BK channel (21). Co-expression of BK channel β4 subunit and α subunit can increase the sensitivity of Martentoxin to BK channel (20).
Fourteen short-chain polypeptides (BmTX1-3, BmKTX, BMP01-03 and Martentoxins) have been isolated and purified from the crude toxin of Buthus Martensi Karsch (BmK) (20). They mainly interaction with BK channel and Small conductance calcium activates potassium channels (SK channel). According to Kalium database (https://kaliumdb.org/), they belong to different α-KTx subfamily. BmTX1-2 belong to the α-KTx 1 subfamily (22,23), BmKTX belong to the α-KTx 3 subfamily, BMP01 belongs to the α-KTx 8 subfamily, BMP02-3 belong to the α-KTx 9 subfamily, BmTX3 belong to the α-KTx 15 subfamily, and martentoxin belongs to the α-KTx 16 subfamily (20). The secondary structure of the α-KTx family is highly conserved and is mostly composed of 23–41 amino acid residues, generally containing 3 pairs of disulfide bonds, 2–3 antiparallel β-folds, and an α-helix (24). However, the sequence similarity of α-KTx family members is low. Studies have shown that changing the disulfide bond pairing form of α-KTx toxin can affect the interaction between the toxin and the targeted potassium channel subtypes (25). Low sequence similarity may be the main factor for the difference in target potassium channel subtypes selection among members of the α-KTx family.
Martentoxin is one of short-chain α-KTx toxins, composed of 37 amino acids and has a molecular weight of 4,060 Da. This compound can specifically inhibit the neuronal BK channel subtype (a+β4) (26). Electrophysiological experiments previously confirmed that the current of the BK channel, which is not sensitive to Iberiotoxin (IbTx), can be blocked by martentoxin, but other types of BK channels are not sensitive to martentoxin (27). The characteristics of the martentoxin-specific interaction with the neuronal BK channel subtype make it a unique ligand for studying the structure and function of the neuronal BK channel subtype. In this work, the mechanism of interaction between Martentoxin and BK channels has been investigated by protein-protein docking, molecular dynamic simulation and virtual alanine mutation methods.
We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6967/rc).
Methods
Homology modeling
Discovery Studio 4.1 [2014] was used for homology modeling of the BK channel (28,29). Position-Specific Iterative (PSI)-BLAST (30,31) was carried out to search for the homology of the BK channel. A potassium channel from Streptomyces lividans [KcsA, Protein Data Bank (PDB) ID 1BL8; crystal structure resolved at 3.2 Å] was selected as a template to build the model on the basis of high similarity and identity with the BK pore (identities =21%, positives =41%). The amino acid sequence of the BK pore was aligned with the sequence of KcsA by Align Sequences in DS 4.1. The Build Homology Model was used to create the homology modeling of the BK channel. Align Sequences and Build Homology Model are the program names for homology modeling in DS software. Refine loops were set at 500 steps, and other parameters were set to default. A total of 420 models were produced. The discrete optimized potential energy (DOPE) score and probability density function (PDF) total energy were employed to evaluate the model. The second homology model was selected for analysis in the follow-up study.
Protein-protein interactions
The crystal structure of martentoxin was found in the Protein Data Bank (PDB ID: 1M2S) (32) and was used in protein-protein docking. A Fast Fourier Transform Correlation technique based protein-protein docking algorithm named ZDOCK was used to establish the BK channel-martentoxin complex (33). In order to get a finer conformational sampling and reach a more accurate prediction, a proper angular step size was set for the sampling of ligand orientations (6° in this study). The top 2,000 poses were evaluated via ZRANK, a widely used scoring function (34). The remainders were analyzed by means of cluster analysis. Next, another algorithm used for the refinement of docked complexes called RDOCK was used to optimize and rerank the docking poses, and to select the best structures that were most consistent with the natural conformation (35). The docked poses were typed with the CHARMm Polar H force field under the default parameters in advance. Finally, the best BK channel-martentoxin complex was used for analysis and other experiments.
Molecular dynamics simulation
The best BK channel-martentoxin complex was prepared by Prepare Protein. Then, the charm force field was added to the compound. Standard Dynamics Cascade was applied for the molecular dynamics (MD) simulations. Prepare Protein and Standard Dynamics Cascade are the names of pre-processing programs for molecular Dynamics simulation performed by DS software. Next, 1,000-step steepest descent minimization and 2,000-step adopted basis NR were performed. Then, the system was heated from 50 K to 300 K in 50 ps and maintained at 300 K for 500 ps to equilibrate under isothermal-isobaric conditions. The conformation was saved every 2 ps and finally a total of 2,500 conformations were obtained.
Virtual alanine mutation scanning (VAMS)
In DS 4.1, VAMS was analyzed by the “Calculate Mutation Energy” protocol, which analyzed the effect of the binding site on BK channel-martentoxin complex binding by a single alanine mutation. The binding energies before and after mutation were calculated by CHARMm using the generalized Born implicit solvent model, which contained empirically scaled contributions of van der Waals, electrostatic interactions and a nonpolar solvation energy term. The difference of binding free energy between mutant structure and wild-type protein was the energetic effect of each mutation on the binding affinity. By cluster analysis of MD simulation trajectory, the input structure of BK-martentoxin complex was obtained. All the residues of martentoxin were selected for calculation. The other parameters were set as default.
Statistical analysis
DS 4.1 and R 3.6.0 software were used to analyze molecular dynamics data and virtual amino acid mutation data. Data are presented as the mean ± standard deviation (SD).
Results
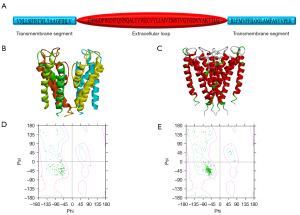
The pore area of the BK channel was composed of 4 identical domains that contained two transmembrane domains and an extracellular loop (Figure 1A). The crystal structure of KcsA from the PDB was used as a template to build the homology pore area model of the BK channel. KcsA has better similarity and identity (identities =21%, positives =41%), so the homology pore area model of the BK channel was built based on the crystal structure of KcsA to ensure good quality and reliability in order to allow structural and functional research. The best model was determined by the lowest value of the probability density function (PDF) total energy and the lowest discrete optimized potential energy (DOPE) score. The Minimization tool and Loop Refinement tool were used to refine the homology model of the BK pore. The best model after repeated optimization is shown in Figure 1B. The 3-dimensional alignment revealed that the BK channel model was not significantly different to the template in terms of the peptide backbone (Figure 1C). The homology model of the BK pore score resulting from the Ramachandran plot in DS 4.1 was also used to evaluate the reliability of the homology model. The Ramachandran plot showed that compared with the amino acid residues of KcsA (Figure 1D), the amino acid residues in the homology model of BK channel were more reasonable (Figure 1E).
Protein-protein docking
To research the interaction mechanism between martentoxin and the pore area of the BK channel, protein-protein docking, a method for obtaining structural information on protein-protein interaction, was employed. The structure of martentoxin was downloaded from PDB (PDB ID: 1M2S, crystal structure resolved at 3.2 Å). The structure of the BK channel was obtained from homology modeling. The model of the BK channel-martentoxin complex was built by the ZDOCK tool in DS 4.1. Then, RDOCK was used to refine all of the poses that were produced by ZDOCK. The top 20 poses were projected to visual analysis. Pose 55 (Figure 2A,2B) was finally selected to analyze the mechanism underlying the interaction of martentoxin with the BK channel. The ZDOCK results showed that hydrogen bonds constituted the important interactions between the BK pores and martentoxin. There were 17 hydrogen bonds revealed from ZDOCK analysis. Phe1 in martentoxin formed 3 hydrogen bonds, and 1 Protein Isoelectric Point (34) interacted with the BK channel, Asn31 also formed 3 hydrogen bonds with the BK channel, Asn32 and Lys28 formed 2 hydrogen bonds with the BK channel, and Lys28 formed a salt bridge interaction with the BK channel (Figure 2C, Table 1). The results show that Phe1, Asn31, Asn32, and Lys28 may be important in the binding of martentoxin to the BK channel.
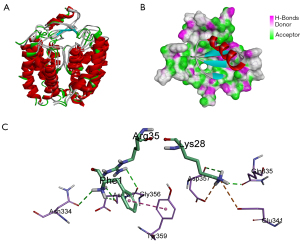
Table 1
| Amino acid name | Bond type | Distance |
|---|---|---|
| Phe1 | Hydrogen | 2.6747 |
| Phe1 | Hydrogen | 2.3114 |
| Phe1 | Hydrogen | 2.8887 |
| Lys7 | Hydrogen | 3.7719 |
| Ser12 | Hydrogen | 3.2815 |
| Trp15 | Hydrogen | 2.8192 |
| Lys28 | Hydrogen | 2.8139 |
| Lys28 | Hydrogen | 2.0233 |
| Lys28 | Salt bridge | 2.9114 |
| Cys29 | Hydrogen | 3.0845 |
| Gln30 | Hydrogen | 2.7725 |
| Asn31 | Hydrogen | 2.9037 |
| Asn31 | Hydrogen | 2.4088 |
| Asn31 | Hydrogen | 3.1408 |
| Asn32 | Hydrogen | 2.6675 |
| Asn32 | Hydrogen | 2.3031 |
| Gln33 | Hydrogen | 2.5837 |
| Arg35 | Hydrogen | 3.0135 |
Arg, Arginine; Asn, Asparagine; Cys, Cysteine; Gln, Glutamine; Leu, Leucine; Lys, Lysine; Phe, Phenylalanine; SER, Serine; Tyr, Tyrosine.
Molecular dynamic simulation and virtual alanine mutation
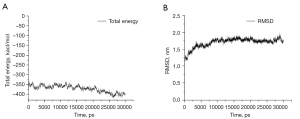
To obtain a more native-like complex structure, we simulated the BK channel-martentoxin complex from the ZDOCK results for 30 ns. The total energy of the system was constant during the MD simulation (Figure 3A). The total energy value suggested that the structure of the BK channel-martentoxin complex was in a dynamic equilibrium state after the equilibrium step was competed. The root mean square deviation (RMSD) of the backbone atoms of the BK channel-martentoxin complex compared to the first structure is shown in Figure 3B. The RMSD values indicate that the system tends to stabilize after 8 ns, and the RMSD value is below 2.00, indicating that the structure of the BK channel-martentoxin complex has good protein stability. When observing the MD simulation in its entirety, we found that the structure of the interaction between the BK channel and martentoxin to not be significantly changed. Some representative structures chosen from the MD simulation had a similar binding pattern. Compared to the first structure, the BK channel-martentoxin complex had more hydrogen bonds and more salt bridges, which improved the binding affinity (Figure 4). The martentoxin residues Phe1, Lys28 and Arg35 formed multiple hydrogen bonds and salt bridges with Asp357, Gly367 and Tyr359 of the BK channel.
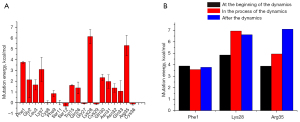
VAMS was performed on the binding residues of martentoxin to evaluate the energetic effect of each residue on the binding affinity. Some typical conformations of BK channel-martentoxin complexes selected from MD simulations are shown in Figure 4. These structures were used to perform VAMS. VAMS of martentoxin revealed that Phe1, which formed 3 hydrogen bonds with the BK channel, is important for binding. Lys28, which was found to form 2 hydrogen bonds and a salt bridge with the BK channel, is also important for binding. Furthermore, although Asn31 formed 3 hydrogen bonds with the BK channel, the mutation of Asn31 had no significant effect on the interaction between the BK channel and martentoxin. In contrast, Arg35 had only 1 hydrogen bond with the BK channel, but the mutation of Arg35 significantly destabilized the binding. Based on this result, we can speculate that the hydrogen bond formed by Arg35 plays an important role in the interaction between the BK channel and martentoxin.
Overall, MD simulation and VAMS were combined to study the mechanism and spatial structure of the interaction between the BK channel and martentoxin. There was a stable interaction between Phe1 and the BK channel in the MD simulation (Figure 4). The interaction between Lys28, Arg35, and the BK channel was enhanced as the MD simulation proceeded (Figure 4). This result may indicate that Phe1 plays an important role in the recognition and binding of martentoxin and the BK channel while Lys28 and Arg35 contribute to the dynamic stability of the BK channel-martentoxin complex.
Discussion
In the nervous system, BK channels control the production of action potentials and the release of transmitters, and functional changes in BK channels lead to a variety of nervous system diseases (36). Results from gene analyses of patients with epilepsy have shown that the mutations of the BK channel protein sequence in patients lead to a prolonged BK channel opening time, increased opening probability, and the subsequent hyperexcitation of neurons, resulting in epilepsy (10). Martentoxin is a specific inhibitor of the BK channel, and thus understanding the interaction mechanism of martentoxin with the BK channel may provide valuable insights into the nature of BK channel inhibitors. The analysis of membrane protein structures has long been a challenging undertaking in the field of protein structure research; however, with the progress of computational biology, homologous modeling is being more widely used in the analysis of membrane protein and ligand interactions (37). In this study, various computational methods were employed to analyze the key amino acid sites of interaction between martentoxin and the BK channel.
The homologous modeling method was performed to construct the structure of the BK channel. Profile-3D and Ramachandran plots were used to analyze the homologous model of the BK channel, and the model of the BK channel was further optimized using the program RDOCK. This ensured that the 3D structure of the BK channel pore area model and the amino acid side chain of the BK channel were within a reasonable range. The BK channel-martentoxin complex was constructed and optimized using ZDOCK and RDOCK, and MD analysis and virtual amino acid mutation analysis were performed. According to the docking results of ZDOCK, it was speculated that the side-chain benzene ring of Phe1 on martentoxin first entered the BK channel pore region and formed electrostatic interactions with amino acids in the BK access pore region. Subsequently, the positively charged amino acids, Lys28 and Arg35, formed electrostatic interactions with negatively charged amino acids in the BK pore region. The subsequent MD results showed that Phe1, Lys28, and Arg35 all had stable interactions with the BK pore region throughout the whole MD simulation. The virtual amino acid mutations also showed a significant increase in BK-martentoxin complex energy after Phe1, Lys28, and Arg35 were mutated to alanine, suggesting that Phe1, Lys28, and Arg35 play important roles in the interaction of martentoxin with the BK pore area.
In some tissues, α subunit and β1-β4 subunits form a variety of subtypes of BK channels. Studies have shown that β1 subunit can improve the binding rate of ChTX to BK channel (21), and the opening rate and opening time of BK channel can be improved after the deglycosylation of β1 (38). The presence of β4 subunit can inhibit the effect of IbTX on BK channel, whereas it can promote the binding of Martentoxin to the BK channel, suggesting that there may be Martentoxin binding sites on the β4 subunit. It is speculated that Martentoxin first interacts with the loop region of the β4 subunit to expose the pore-area binding site of BK channel, then Martentoxin interacts with the pore-area binding site of the BK channel. Due to the large spatial flexibility of β4 loop region, the spatial structure of the loop region of the β4 subunit is difficult be accurately analyzed in MD simulation, the function of the β subunit in the ligand-BK channel interaction species can be further studied by molecular biology and electrophysiological techniques. In the year 2020, the molecular mechanism underlying the interaction of β4 subunit with Martentoxin has been reported. Researchers using a variety of methods have shown that a series of specific amino acid residues played a key role in the interaction between the toxin and channel, including the residues Asp5, Glu13, Lys20, Ser24, Gln26, Lys28, and Arg35 of the martentoxin, and the residues Glu104, Glu122, Gln124, Lys125, and Glu128 in the hβ4-loop of BK channel. These key amino acids between the toxin and the channel greatly enhance the toxin-channel binding ability by forming hydrogen bonds (39). The identification of important martentoxin amino acids that interact with BK can provide a basis for the study of BK channel-specific inhibitors. In the future, we are going to use more tools, including amino acids site-directed mutation and electrophysiological techniques, to further analyze the key amino acids of the interaction between martensite toxin and BK channel.
Acknowledgments
Funding: This research was supported by National Natural Science Foundation of China (31771191), partly funded by Chongming Science and Technology Committee (CKY2019-06, CKY2021-38) and Xinhua Hospital Chongming Branch Scientific Program (2019YA03, 2019YA06).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6967/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6967/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6967/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Brandalise F, Ratto D, Leone R, et al. Deeper and Deeper on the Role of BK and Kir4.1 Channels in Glioblastoma Invasiveness: A Novel Summative Mechanism? Front Neurosci 2020;14:595664. [Crossref] [PubMed]
- Marty A. Ca-dependent K channels with large unitary conductance in chromaffin cell membranes. Nature 1981;291:497-500. [Crossref] [PubMed]
- Vicens A, Vinuesa P, Arenas M, et al. Analyzing the functional divergence of Slo1 and Slo3 channel subfamilies. Mol Phylogenet Evol 2019;133:33-41. [Crossref] [PubMed]
- Gonzalez-Perez V, Lingle CJ. Regulation of BK Channels by Beta and Gamma Subunits. Annu Rev Physiol 2019;81:113-37. [Crossref] [PubMed]
- Ishii TM, Silvia C, Hirschberg B, et al. A human intermediate conductance calcium-activated potassium channel. Proc Natl Acad Sci U S A 1997;94:11651-6. [Crossref] [PubMed]
- Ning FL, Tao J, Li DD, et al. Activating BK channels ameliorates vascular smooth muscle calcification through Akt signaling. Acta Pharmacol Sin 2021; [Epub ahead of print]. [Crossref] [PubMed]
- Wan XJ, Zhao HC, Zhang P, et al. Involvement of BK channel in differentiation of vascular smooth muscle cells induced by mechanical stretch. Int J Biochem Cell Biol 2015;59:21-9. [Crossref] [PubMed]
- He C, Li X, Wang M, et al. Deletion of BK channels decreased skeletal and cardiac muscle function but increased smooth muscle contraction in rats. Biochem Biophys Res Commun 2021;570:8-14. [Crossref] [PubMed]
- Rüttiger L, Sausbier M, Zimmermann U, et al. Deletion of the Ca2+-activated potassium (BK) alpha-subunit but not the BKbeta1-subunit leads to progressive hearing loss. Proc Natl Acad Sci U S A 2004;101:12922-7. [Crossref] [PubMed]
- Li X, Poschmann S, Chen Q, et al. De novo BK channel variant causes epilepsy by affecting voltage gating but not Ca2+ sensitivity. Eur J Hum Genet 2018;26:220-9. [Crossref] [PubMed]
- Yang J, Krishnamoorthy G, Saxena A, et al. An epilepsy/dyskinesia-associated mutation enhances BK channel activation by potentiating Ca2+ sensing. Neuron 2010;66:871-83. [Crossref] [PubMed]
- Bailey CS, Moldenhauer HJ, Park SM, et al. KCNMA1-linked channelopathy. J Gen Physiol 2019;151:1173-89. [Crossref] [PubMed]
- Chen X, Wang Q, Ni F, et al. Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement. Proc Natl Acad Sci U S A 2010;107:11352-7. [Crossref] [PubMed]
- Doyle DA, Morais Cabral J, Pfuetzner RA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 1998;280:69-77. [Crossref] [PubMed]
- McManus OB, Harris GH, Giangiacomo KM, et al. An activator of calcium-dependent potassium channels isolated from a medicinal herb. Biochemistry 1993;32:6128-33. [Crossref] [PubMed]
- Morimoto T, Sakamoto K, Sade H, et al. Voltage-sensitive oxonol dyes are novel large-conductance Ca2+-activated K+ channel activators selective for beta1 and beta4 but not for beta2 subunits. Mol Pharmacol 2007;71:1075-88. [Crossref] [PubMed]
- Imaizumi Y, Sakamoto K, Yamada A, et al. Molecular basis of pimarane compounds as novel activators of large-conductance Ca(2+)-activated K(+) channel alpha-subunit. Mol Pharmacol 2002;62:836-46. [Crossref] [PubMed]
- Ahn HS, dela Peña I, Kim YC, et al. 4-Chloro-7-trifluoromethyl-10H- benzo4,5furo3,2-bindole-1-carboxylic acid (TBIC), a putative BK(Ca) channel opener with uterine relaxant activities. Pharmacology 2011;87:331-40. [Crossref] [PubMed]
- Ghatta S, Nimmagadda D, Xu X, et al. Large-conductance, calcium-activated potassium channels: structural and functional implications. Pharmacol Ther 2006;110:103-16. [Crossref] [PubMed]
- Tao J, Zhou ZL, Wu B, et al. Recombinant expression and functional characterization of martentoxin: a selective inhibitor for BK channel (α + β4). Toxins (Basel) 2014;6:1419-33. [Crossref] [PubMed]
- Hanner M, Schmalhofer WA, Munujos P, et al. The beta subunit of the high-conductance calcium-activated potassium channel contributes to the high-affinity receptor for charybdotoxin. Proc Natl Acad Sci U S A 1997;94:2853-8. [Crossref] [PubMed]
- Gao B, Peigneur S, Dalziel J, et al. Molecular divergence of two orthologous scorpion toxins affecting potassium channels. Comp Biochem Physiol A Mol Integr Physiol 2011;159:313-21. [Crossref] [PubMed]
- Xu CQ, Brône B, Wicher D, et al. BmBKTx1, a novel Ca2+-activated K+ channel blocker purified from the Asian scorpion Buthus martensi Karsch. J Biol Chem 2004;279:34562-9. [Crossref] [PubMed]
- Tabakmakher VM, Krylov NA, Kuzmenkov AI, et al. Kalium 2.0, a comprehensive database of polypeptide ligands of potassium channels. Sci Data 2019;6:73. [Crossref] [PubMed]
- M'Barek S, Lopez-Gonzalez I, Andreotti N, et al. A maurotoxin with constrained standard disulfide bridging: innovative strategy of chemical synthesis, pharmacology, and docking on K+ channels. J Biol Chem 2003;278:31095-104. [Crossref] [PubMed]
- Shi J, He HQ, Zhao R, et al. Inhibition of martentoxin on neuronal BK channel subtype (alpha+beta4): implications for a novel interaction model. Biophys J 2008;94:3706-13. [Crossref] [PubMed]
- Tao J, Shi J, Yan L, et al. Enhancement effects of martentoxin on glioma BK channel and BK channel (α+β1) subtypes. PLoS One 2011;6:e15896. [Crossref] [PubMed]
- Gan G, Yi H, Chen M, et al. Structural basis for toxin resistance of beta4-associated calcium-activated potassium (BK) channels. J Biol Chem 2008;283:24177-84. [Crossref] [PubMed]
- Jiang ZY, Chu HX, Xi MY, et al. Insight into the intermolecular recognition mechanism between Keap1 and IKKβ combining homology modelling, protein-protein docking, molecular dynamics simulations and virtual alanine mutation. PLoS One 2013;8:e75076. [Crossref] [PubMed]
- Ding S, Li Y, Shi Z, et al. A protein structural classes prediction method based on predicted secondary structure and PSI-BLAST profile. Biochimie 2014;97:60-5. [Crossref] [PubMed]
- Bhagwat M, Aravind L. PSI-BLAST tutorial. Methods Mol Biol 2007;395:177-86. [Crossref] [PubMed]
- Wang Y, Chen X, Zhang N, et al. The solution structure of BmTx3B, a member of the scorpion toxin subfamily alpha-KTx 16. Proteins 2005;58:489-97. [Crossref] [PubMed]
- Chen R, Weng Z. Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins 2002;47:281-94. [Crossref] [PubMed]
- Pierce B, Weng Z. ZRANK: reranking protein docking predictions with an optimized energy function. Proteins 2007;67:1078-86. [Crossref] [PubMed]
- Li L, Chen R, Weng Z. RDOCK: refinement of rigid-body protein docking predictions. Proteins 2003;53:693-707. [Crossref] [PubMed]
- Kim Y, Park J, Choi YK. The Role of Astrocytes in the Central Nervous System Focused on BK Channel and Heme Oxygenase Metabolites: A Review. Antioxidants (Basel) 2019;8:121. [Crossref] [PubMed]
- Al-Obaidi A, Elmezayen AD, Yelekçi K. Homology modeling of human GABA-AT and devise some novel and potent inhibitors via computer-aided drug design techniques. J Biomol Struct Dyn 2021;39:4100-10. [Crossref] [PubMed]
- Hagen BM, Sanders KM. Deglycosylation of the beta1-subunit of the BK channel changes its biophysical properties. Am J Physiol Cell Physiol 2006;291:C750-6. [Crossref] [PubMed]
- Liu X, Tao J, Zhang S, et al. Selective Blockade of Neuronal BK (α + β4) Channels Preventing Epileptic Seizure. J Med Chem 2020;63:216-30. [Crossref] [PubMed]
(English Language Editor: J. Gray)


