
Clinical and molecular features of sacrum chordoma in Chinese patients
Introduction
Chordoma is a rare bone tumor of the axial skeleton that arises from remnants of the notochord. Most chordomas occur in the skull base, sacrum, or mobile spine, and usually occur in people aged 50–60 years old (1). The mainstay treatment for chordoma is complete surgical resection; radiation after surgery is also necessary for patient whose focus is difficult to remove by surgery entirely, around which there are many important vessels and nerves, was difficult to entirely remove by surgery. Because of these vital structures, chordoma has high recurrence and metastasis rates (26.7–66.7%) that result in a poor prognosis (2). Compared with common malignant bone tumors, chordoma has a relatively low degree of malignancy and slow growth. Chordoma usually has no typical clinical symptoms in the early stage and is often diagnosed in the late stage, invading the important anatomical structures around it. As only limited treatment options are available, there is an urgent need to explore effective therapeutic strategies to improve chordoma outcomes and survival rates.
A large number of studies have shown that the T-box transcription factor T (TBXT) gene is a key gene in chordoma, but the expression of its coded protein, brachyury, is not related to the prognosis of chordoma, and currently, it can only be used as a diagnostic marker gene (3). Previous studies have shown that deoxyribonucleic acid (DNA) copy number alternations (CNAs), such as Cyclin Dependent Kinase Inhibitor 2A (CDKN2A), Phosphatase And Tensin Homolog (PTEN), and SWItch/Sucrose Nonfermentable Related, Matrix Associated, Actin Dependent Regulator Of Chromatin, Subfamily B, Member 1 (SMARCB1), play key roles in the development of sacrum chordoma (4). A recent research study based on the whole-exome-sequencing (WES) and whole-genome-sequencing (WGS) of chordomas (including 48 sacrum chordoma cases) observed recurrent mutations in PI3K signaling genes, including PIK3CA, PIK3R1 and PTEN, chromatin modelling genes, including AT-Rich Interaction Domain 1A (ARID1A), Polybromo 1 (PBRM1) and SET Domain Containing 2, Histone Lysine Methyltransferase (SETD2), and Lysosomal Trafficking Regulator (LYST) gene (5).
With the development of next-generation sequencing technology, novel somatic mutations of sacrum chordoma may be able to be identified and new targeted drug therapies developed. The exon region is an important region in the genome that encodes proteins, accounting for about 1% of the genome, but it contains about 85% of the pathogenic mutations. WES is a genomic analysis method that uses sequence capture technology to capture and enrich the DNA sequence of the exon region of the genome for high-throughput sequencing. Compared with WGS, WES is more economical and efficient and has a greater advantage in the study of single nucleotide polymorphism (SNP) and insertions/deletion (InDel) and a higher sequencing depth which can find mutations that WGS cannot find (6,7). In recent years, WES has successfully identified related mutations in solid tumors such as leukemia, myeloma and renal cell carcinoma (8,9). Eight patients with complete clinical data were selected for the study. In this study, we performed WES on 8 sacrum chordoma tissue samples and matched peripheral blood samples from patients. After comparing the tumor tissue with the blood tissue, we excluded the germline mutations and obtained the somatic mutations which were analyzed from the aspects of driver genes, pathways enrichment and SMGs. The results of the analysis were verified from the aspects of gene sequence and protein expression. We obtained the gene mutational signatures of sacrum chordoma in Chinese patients and identified novel driver genes and pathways closely linked to the progress of this incurable disease.
We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6617/rc).
Methods
Samples and clinical data
Tumor tissues and blood were obtained from 8 patients who underwent sacrum tumor resection surgery at the Department of Orthopedics of The First Affiliated Hospital of Soochow University. The tumor tissues were examined by the Pathology Department, and blood samples were drawn from each patient’s upper arm vein before surgery. For detailed information on the 8 patients, see Table 1.
Table 1
| Characteristic | Selected cases |
|---|---|
| Total | 8 |
| Median age (range), years | 57.5 [36–71] |
| Gender, No. (%) | |
| Male | 5 (62.5) |
| Female | 3 (37.5) |
| Primary/recurring | 4/4 (1:1) |
| Pre-surgery ECOG score, No. (%) | |
| 0 | 2 (25.0) |
| 1 | 6 (75.0) |
| 2 | 0 |
| Post-surgery ECOG score, No. (%) | |
| 0 | 1 (12.5) |
| 1 | 2 (25.0) |
| 2 | 4 (50.0) |
| 3 | 1 (12.5) |
| Immunohistochemistry expressed proteins, % (No.) | |
| S100 | 75.00% (6/8) |
| CK | 100.00% (8/8) |
| Vimentin | 100.00% (8/8) |
| Ki-67 | 100.00% (8/8) |
| EMA | 100.00% (7/7) |
| CK18 | 100.00% (8/8) |
| Ki-67 % | 4 (1–15%) |
| Antithrombin III activity % (range) | 92.5 (67–118%) |
ECOG, Eastern Cooperative Oncology Group; S100, S100 Calcium Binding Protein; CK, Cytokeratin; Ki-67, Proliferation Marker Protein KI-67; EMA, epithelial membrane antigen; CK18, Cytokeratin 18.
All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of The First Affiliated Hospital of Soochow University (2021 No.341) and informed consent was taken from all the patients.
WES
Genomic DNA were extracted from the tumor samples and matched peripheral blood and stored in liquid nitrogen according to the standard protocols. The qualified DNAs were randomly fragmented into 150–220 bp by Covaries Sample Preparation System. The exome library was constructed using the Agilent SureSelect Human All Exon V6/V7, and the captured DNA library was sequenced on the Illumina HiSeq X Ten PE150 platform. The clean reads acquired using FASTP were mapped to the human_glk_v37 reference genome using Burrows-Wheeler Aligner (10). The duplicate reads, which were produced by polymerase chain reaction (PCR), were removed with a Picard after converting the format using Sequence Alignment/Map (SAM) tools (11). The average sequencing coverage achieved was 136 X in the tumor samples and 139 X in the blood samples. The Genome Analysis Toolkit (GATK4) was used to identify SNPs and INDELs (12). The test results were annotated to a number of databases, including the Reference Sequence database (Refseq), 1000Genoms Project, the Exome Aggregation Consortium (EXAC), NHLBI GO Exome Sequencing Project6500 (ESP6500), Sorting Intolerant From Tolerant) SIFT, PolyPhen, Catalogue Of Somatic Mutations In Cancer) COSMIC databases, and Annovar software was used to identify the candidate SNPs or INDELs (13,14). To identify the somatic mutations, the tumor tissues were aligned to the blood samples. By comparing the tumor tissues with the peripheral blood samples, we filtered out the germline mutations in the blood samples and retained only the somatic mutations identified in the tumor cells during the analysis.
Driver genes and pathways
We compared the somatic mutations to the known driver genes, which were recorded in Cancer Gene Census, Bert Volelstein125 (15), and 127 significantly mutated gene in The Cancer Genome Atlas (TCGA) pan-cancer (SMG127) (16), to screen the driver mutations in the tumor samples. We used the PathScan of Mutational Significance in Cancer (MuSiC) software in the enrichment pathways analysis to identify the significantly altered pathways in which the somatic mutations participated (17). The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used in the analysis.
Significantly mutated genes (SMGs)
To identify the SMGs, we used MuSiC software to identify the genes whose mutation rate was significantly higher than the background mutation rate in the somatic mutations of all the tumor samples.
Immunohistochemistry
The tumor tissues were fixed in 10% formalin for 6 hours and embedded in paraffin. Conventional slices (4 µm) were harvested on glass slides pre-treated with 2% (3-aminopropyltriethoxysilane) APES acetone and dried in a 60 °C oven for 1–2 hours. Immunohistochemistry staining was performed on the slices using the BenchMark XT automatic multi-function histopathological detection system for the following antibodies: CDKN2A (SC-1661, 1:200; Santa Cruz Biotechnology, Inc.), Calreticulin (CALR) (SC-373863, 1:200; Santa Cruz Biotechnology, Inc.), ROS Proto-Oncogene 1, Receptor Tyrosine Kinase (ROS1) (SC-376217, 1:100; Santa Cruz Biotechnology, Inc.), PIK3CA (SC-8010, 1:100; Santa Cruz Biotechnology, INC), Transcription Factor EB (TFEB) (SC-166736, 1:100; Santa Cruz Biotechnology, Inc.), TBXT (SC-166962, 1:50; Santa Cruz Biotechnology, Inc.), Chromodomain Helicase DNA Binding Protein 3 (CHD3) (SC-55606, 1:50; Santa Cruz Biotechnology, Inc.), Cut Like Homeobox 1 (CUX1) (SC-514008, 1:50; Santa Cruz Biotechnology, Inc.). The positive controls and negative controls of each antibody showed the appropriate results.
Sanger sequencing
Reverse transcription PCR (RT-PCR) was performed on 1 µg of the total ribonucleic acid (RNA) to validate the SMGs. The PCR primers were designed by Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA, USA). All the amplifications underwent the following identical cycling profiles: an initial denaturation at 95 °C for 3 min, followed by 35 cycles of 30 sec at 95 °C, 30 sec at 58 °C, and 30 sec at 72 °C, and a final extension for 7 min at 72 °C annealing. The RT-PCR-amplified products were purified using a QIAGEN gel extraction kit (Qiagen, Hilden, Germany), and then sequenced using the Big Dye Terminator cycle sequencing kit (Applied Biosystems, USA) under the following conditions: initial denaturation at 96 °C for 1 min and 25 cycles of 10 sec at 96 °C, 5 sec at 50 °C, and 4 min at 60 °C.
GeneChip
The genomic DNA was digested, ligated, amplified, fragmented, and labeled using a Cytoscan Optima kit (Thermo Fisher Company, USA), and the product was hybridized with a Cytoscan Optima chip (containing 315,000 probes) overnight. The chip was washed and dyed by a washing station (450Dxv.2, Thermo Fisher, USA), and the data were collected by the Affymetrix 3000Dxv.2 chip scanner. ChAS software was used to interpret the results.
Statistical analysis
Statistical analyses were performed using SPSS version 25.0 (SPSS, Inc., Chicago, IL, USA). Data were analyzed by the Fisher exact tests, convolution test and likelihood ratio test. P<0.05 was considered to a statistically significant difference.
Results
Patient characteristics
From 1996 to 2019, our Department in China treated and followed up with 50 sacrum chordoma patients who underwent targeted vessel embolization. We performed WES on 8 sacrum chordoma tissues (4 primary tumors and 4 recurrences), all of which had matching peripheral blood samples that had been drawn before surgery. Their clinical characteristics of the patients are shown in Table 1.
Somatic genomic alterations of sacrum chordoma
WES was performed on the genomic DNA of the sacrum chordoma specimens and their matched blood specimens. Somatic alterations, including non-synonymous mutations (see Figure 1A) and CNAs (see Figure 1B), were identified. The somatic alterations among these patients were very different. Patients #1 and #2 had the most abundant non-synonymous mutations (459 and 2,509, respectively). The other patients had non-synonymous mutations ranging from 4 to 64. Patients #3, #4, and #6 had the most abundant genes with CNAs (>1,000). Patient #8 had the least number of non-synonymous mutations and the least number of CNAs.
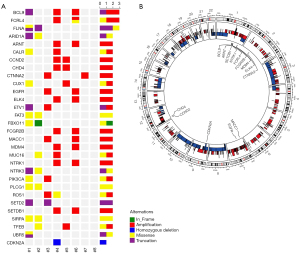
To identify the potential driver genes in sacrum chordoma, we analyzed the cancer-related genes reported in the Cancer Gene Census, Bert Vogelstein 125 (15), and SMG 127 (16). In relation to the CNAs, only oncogenes with amplification and tumor suppressor genes with homozygous deletion were included in the genomic profiling (see Figure 1A). The most frequently altered gene was B-Cell Lymphoma 9 Protein (BCL9), which was altered in 3 patients. The other genes altered in more than 1 patient were ARID1A, epidermal growth factor receptor (EGFR), Neurotrophic Receptor Tyrosine Kinase 1/3 (NTRK1/3), PIK3CA, and SETD2, but many other cancer-related genes were also identified (see Figure 1A). The homozygous deletion of CDKN2A, which occurs frequently in other chordoma cohorts (5), was only found in 1 patient in our cohort (see Figure 1A). In 1 of the 8 tumor genomes, we found the common nonsynonymous SNP rs2305089 of the TBXT gene, which is strongly associated with chordoma (18). The Variant allele Frequency (VAF) values of all the related genes are shown in the Table S1.
In sacrum chordoma, CNAs could be found in any chromosome (see Figure 1B). The loss of 1 allele was observed in almost the entire chromosomes of 3, 8, 9, 10, 14, and 18. The homozygous deletion of genes was relatively rare in these patients. Amplifications of copy numbers were diffusely distributed in the chromosomes and the copy numbers increased to 3–5 copies.
Mutations in PI3K signaling genes and chromatin modelling genes
Using KEGG, we found that the mutated genes in these sacrum chordoma specimens were enriched in the transcriptional mis-regulation pathway, the PI3K/mTOR signaling pathway, and the extracellular matrix (ECM)-receptor interaction pathway (see Figure 2A), which have been demonstrated to play important roles in cancer development, especially in the promotion of cell proliferation.
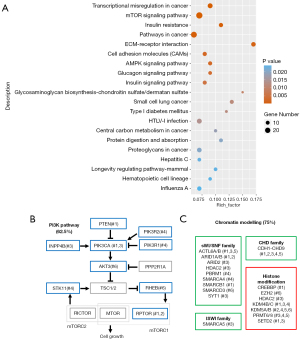
Next, we analyzed the PI3K signaling genes and chromatin modelling genes, which are related to the above-mentioned pathways and have been reported in chordoma (5) and diverse tumor types (19,20). Interestingly, we found that 5 (62.5%) of the 8 patients had at least 1 alteration in genes of the PI3K pathway, including PTEN, PIK3CA, PIK3R1/2, AKT Serine/Threonine Kinase 3 (AKT3), Serine/Threonine Kinase 11 (STK11), Ras Homolog, MTORC1 Binding (RHEB), and Regulatory Associated Protein Of MTOR Complex 1 (RPTOR) (see Figure 2B). Six patients had mutations in 3 subfamilies of the chromatin modelling genes and histone modification genes. These mutated genes included the SWItch/Sucrose Nonfermentable (SWI/SNF) family genes ARID1A, Histone Deacetylase 2 (HDAC2), PBRM1, the imitation switch (ISWI) family gene SMARCA5, the CHD family genes of CHD1 to CHD9, and the histone modification genes of Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit (EZH2) and SETD2 (see Figure 2C).
CLDN9 as a potential oncogene in sacrum chordoma
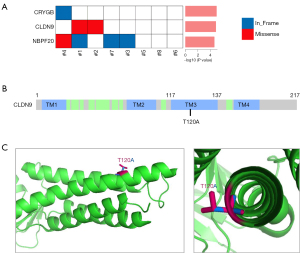
To identify the potential driver events in tumorigenesis, we conducted a MuSiC (21) analysis of the somatic mutations of all the tumor samples and discovered 3 SMGs; that is, Crystallin Gamma B (CRYGB), recombinant CLDN9, and Neuroblastoma Breakpoint Family Member 20 (NBPF20) (see Figure 3A). Mutations in CRYGB were found in 12.5% of the tumor samples, including 2 non-frameshift indels (c.103_108del, p.35_36del; Variant Allen Frequency: 34%, and c.110_115del, p.37_39del; Variant Allen Frequency: 35%). The recurrent mutation in CLDN9 was found in 25% of the tumor samples, including 1 missense mutation (c.A358G, p.T120A; Variant Allen Frequency: 7%; see Figure 3B). Mutations in NBPF20 were found in 50% of the tumor samples, including 1 missense mutation (c.G8194A, p. D2732N; Variant Allen Frequency: 23%) and 1 non-frameshift indel (c.8168_8169insGAG, p. R2723delinsRR; Variant Allen Frequency: 14%).
Verification of related genes
The 3 SMGs were subjected to rigorous validation by Sanger sequencing in all the related samples. The mutations acquired from WES outcomes were compared to those acquired from the Sanger sequencing outcomes in the 3 SMGs to confirm which recurrently mutated genes are involved in chordoma. The CLDN9 and CRYGB mutations had the same mutations at the same point as those found in the WES.
In order to verify the results of WES at the protein level, we selected several relative important genes for immunohistochemistry. The immunohistochemistry analysis revealed that PIK3CA, CALR, CUX1, and ROS1 proteins, which were identified by WES (see Figure 1A) had positive stains in the corresponding tumor samples (see Figure S1A-S1D). CALR, in particular, was positive in all the tumor samples.
A GeneChip assay (see Figure S1E,S1F) was performed on Patient #4, who had a relatively large number of copy number losses/gains, and all the losses/gains identified by the WES were verified (see Figure 1A and https://cdn.amegroups.cn/static/public/atm-21-6617-1.xlsx). Brachyury, which is a decisive marker of chordoma, was also positive in all the tumor samples (see Figure S2).
Discussion
Chordoma arises from remnants of the notochord and was first characterized microscopically by Virchow in 1857 (22). At present, the most convincing hypothesis for the incidence of chordoma is the discovery of a gene duplication in TBXT gene which was expressed in almost all chordomas, but the specific pathogenesis is still unclear. Therefore, it is of great significance to explore the new driver genes and signal pathways of chordoma besides TBXT gene.
Here, we presented a comparatively deep exploration of chordoma patients in China from the perspective of genomics and revealed the cancer genome of chordoma centering on the driver landscape of somatic mutant.
By comparing with KEGG, we found that 75% of the patients had mutations in three subfamilies of chromatin modeling genes and histone modified genes, suggesting that the defect of chromatin modeling genes may be one of the driving forces for the pathogenesis of chordoma. 62.5% of the patients had at least one mutation in the PI3K pathway. PI3K signaling pathway has been proved to play an important role in the occurrence and development of tumors and is a target for a variety of tumor therapies. Our findings also provide a reasonable explanation for its use as a target for chordoma therapy.
CLDN9 is a member of the claudin family that has been reported to participate in the signaling pathways linked to neoplasia and exert a mutual effect on tight junction proteins. The overexpression of CLDN9 is associated with the increased infiltration of pituitary tumors (23), the invasion and metastasis of gastric adenocarcinoma cells in vitro (24), and the metastatic ability of hepatocytes in vitro (25).
However, the CLDN9 T120A substitution has not been previously reported. We used the 3-dimensional structure of CLDN9 (26) to mimic the T120A point mutation and found that it was located in 1 of the transmembrane domains, and amino acid substitution from Threonine (Thr) to Alanine (Ala) lost an alkyl-side-chain outside the helix surface (see Figure 3C). This may affect its interaction with membranes or other proteins.
Our observation of the CALR gene which was all positive in immunohistochemistry shows that CALR, which plays a role in protein folding quality control and calcium homeostasis (27), may play an important role in the development of chordoma. Although this hypothesis needs to be further proved, one can speculate on the biological role of CALR in the pathogenesis of chordoma. CALR has been found in the nucleus, indicating that it may play a role in transcriptional regulation, and recurrent mutations of CALR may be closely related to tumorigenesis.
With the progress of various treatments, the local control rate of chordoma has been effectively improved. However, the long-term survival rate is still very low, with a 5-, 10- and 20-year survival rate of 47–80%, 40% and 13% (2,28). Respectively, the recurrence rate of chordoma is very high and the curative effect of surgery or re-radiotherapy for recurrent patients is very limited. Therefore, as a new direction of tumor therapy, molecular targeted drug therapy has been studied by more and more scholars. Targeted drugs can inhibit a specific molecular site or signal pathway to inhibit the growth of tumor cells and promote apoptosis. In summary, in our study we profiled the genomic alteration of sacrum chordoma in Chinese patients. Our findings could be potential therapeutic molecular targets and used to develop a potential promising therapeutic strategy for the treatment of sacrum chordoma.
Acknowledgments
Funding: This study was supported by grants from the National Key R&D Program of China (No. 2019YFA0111000); the National Natural Science Foundation of China (Nos. 81600116, 81600114, 81700140, 81970142, 81900130, 81970136, and 82000132); the Natural Science Foundation of the Jiangsu Higher Education Institution of China (No. 18KJA320005); the Natural Science Foundation of Jiangsu Province (No. BK20190180); the China Postdoctoral Science Foundation (No. 2018M632372); and the priority academic development program of Jiangsu Higher Education Institution, a Translational Research Grant of National Clinical Research Center for Hematological Diseases (NCRCH) (Nos. 2020WSB11 and 2020WSB13).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6617/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6617/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6617/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of The First Affiliated Hospital of Soochow University (2021 No.341) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- McMaster ML, Goldstein AM, Bromley CM, et al. Chordoma: incidence and survival patterns in the United States, 1973-1995. Cancer Causes Control 2001;12:1-11. [Crossref] [PubMed]
- Sahyouni R, Goshtasbi K, Mahmoodi A, et al. A historical recount of chordoma. J Neurosurg Spine 2018;28:422-8. [Crossref] [PubMed]
- Tirabosco R, Mangham DC, Rosenberg AE, et al. Brachyury expression in extra-axial skeletal and soft tissue chordomas: a marker that distinguishes chordoma from mixed tumor/myoepithelioma/parachordoma in soft tissue. Am J Surg Pathol 2008;32:572-80. [Crossref] [PubMed]
- Choy E, MacConaill LE, Cote GM, et al. Genotyping cancer-associated genes in chordoma identifies mutations in oncogenes and areas of chromosomal loss involving CDKN2A, PTEN, and SMARCB1. PLoS One 2014;9:e101283. [Crossref] [PubMed]
- Tarpey PS, Behjati S, Young MD, et al. The driver landscape of sporadic chordoma. Nat Commun 2017;8:890. [Crossref] [PubMed]
- Hodges E, Xuan Z, Balija V, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet 2007;39:1522-7. [Crossref] [PubMed]
- Ku CS, Wu M, Cooper DN, et al. Exome versus transcriptome sequencing in identifying coding region variants. Expert Rev Mol Diagn 2012;12:241-51. [Crossref] [PubMed]
- Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 2010;42:181-5. [Crossref] [PubMed]
- Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 2010;463:360-3. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078-9. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 2011;39:D945-50. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [Crossref] [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333-9. [Crossref] [PubMed]
- Wendl MC, Wallis JW, Lin L, et al. PathScan: a tool for discerning mutational significance in groups of putative cancer genes. Bioinformatics 2011;27:1595-602. [Crossref] [PubMed]
- Pillay N, Plagnol V, Tarpey PS, et al. A common single-nucleotide variant in T is strongly associated with chordoma. Nat Genet 2012;44:1185-7. [Crossref] [PubMed]
- Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018;173:321-337.e10. [Crossref] [PubMed]
- Chen J, Herlong FH, Stroehlein JR, et al. Mutations of Chromatin Structure Regulating Genes in Human Malignancies. Curr Protein Pept Sci 2016;17:411-37. [Crossref] [PubMed]
- Dees ND, Zhang Q, Kandoth C, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res 2012;22:1589-98. [Crossref] [PubMed]
- Walcott BP, Nahed BV, Mohyeldin A, et al. Chordoma: current concepts, management, and future directions. Lancet Oncol 2012;13:e69-76. [Crossref] [PubMed]
- Hong L, Wu Y, Feng J, et al. Overexpression of the cell adhesion molecule claudin-9 is associated with invasion in pituitary oncocytomas. Hum Pathol 2014;45:2423-9. [Crossref] [PubMed]
- Zavala-Zendejas VE, Torres-Martinez AC, Salas-Morales B, et al. Claudin-6, 7, or 9 overexpression in the human gastric adenocarcinoma cell line AGS increases its invasiveness, migration, and proliferation rate. Cancer Invest 2011;29:1-11. [Crossref] [PubMed]
- Liu H, Wang M, Liang N, et al. Claudin-9 enhances the metastatic potential of hepatocytes via Tyk2/Stat3 signaling. Turk J Gastroenterol 2019;30:722-31. [Crossref] [PubMed]
- Vecchio AJ, Stroud RM. Claudin-9 structures reveal mechanism for toxin-induced gut barrier breakdown. Proc Natl Acad Sci U S A 2019;116:17817-24. [Crossref] [PubMed]
- Klampfl T, Gisslinger H, Harutyunyan AS, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med 2013;369:2379-90. [Crossref] [PubMed]
- Stacchiotti S, Gronchi A, Fossati P, et al. Best practices for the management of local-regional recurrent chordoma: a position paper by the Chordoma Global Consensus Group. Ann Oncol 2017;28:1230-42. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


