
Autophagy related long non-coding RNA and breast cancer prognosis analysis and prognostic risk model establishment
Introduction
The latest cancer reports show that breast cancer (BRCA) has replaced lung cancer as the leading cancer worldwide, posing a serious threat to women’s health (1,2). Alongside important advances in the treatment of BRCA, developments in antitumor therapies have greatly improved the survival rate and quality of life of tumor patients. However, with the use of antineoplastic drugs, resistance gradually develops, leading to reduced therapeutic efficacy, treatment relapse, metastasis, and finally to treatment failure. Although the means of diagnosis and treatment of BRCA have improved, it has maintained a high mortality rate (3,4). Tumor metastasis and recurrence are complex biological processes, and traditional tumor markers carcinoembryonic antigen (CEA) and carbohydrate antigen 153 (CA153) are no longer recommended as indicators for the diagnosis and postoperative follow-up of BRCA because they lack sufficient sensitivity and specificity. The occurrence and progression of BRCA is accompanied by abnormal regulation at multiple genes and levels (5,6). The search for effective diagnostic and prognostic markers is of great importance in the prevention and treatment of BRCA.
In recent years, the rapid development of high-throughput sequencing technology has led to a deeper understanding of molecular biology, especially non-coding RNAs. Long-stranded non-coding RNAs (lncRNAs) are a class of endogenous RNAs with transcripts longer than 200 nucleotides (7). Studies have reported that lncRNAs are involved in tumorigenesis, progression, metastasis, and immune response and are associated with poor prognosis (8,9). Interfering with lncRNA expression can alter the tumor profile and has been widely recognized in several cancer types (10). Recent studies have shown that tissue-specific expression profiles of lncRNAs may represent tissue specificity for different stages of BRCA and may serve as biomarkers or therapeutic targets for BRCA diagnosis and prognosis.
Autophagy is a highly conserved self-degradation mechanism that occurs within eukaryotic cells and is fundamental to the maintenance of organismal health (11). Alterations in autophagy are closely associated with various diseases such as tumors, metabolic disorders, and neurodegeneration (12). An increasing number of studies have shown that lncRNAs are involved in cancer development by regulating autophagy levels and have an important role in prognostic assessment with BRCA (13,14). However, the significance of autophagy-associated lncRNAs in BRCA prognosis has been rarely reported. Identifying autophagy lncRNAs that are closely associated with BRCA and prognosis is of great importance. In this study, we proposed to screen out BRCA autophagy-related lncRNAs and construct a risk scoring model in order to provide a new reference for clinicians to predict the prognosis of BRCA patients. Recent studies have shown that BRCA is not only composed of tumor cells, but also has significant changes in the surrounding stroma or tumor microenvironment. For example, inhibitory immune cells, soluble factors and changed extracellular matrix work together to hinder effective anti-tumor immunity and promote the progression and metastasis of BRCA (15). Herein, we investigated the relationship between autophagy risk model and immune infiltration of BRCA, which laid the foundation for autophagy-related immunotherapy. We present the following article in accordance with the TRIPOD reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6251/rc).
Methods
Raw data
The transcriptomic data of 1,109 BRCA samples and 113 normal samples were downloaded from The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/), along with clinical data of BRCA patients, including age, tumor-node-metastasis (TNM) stage, tumor stage, distant metastasis, overall survival (OS) time, and survival status. We also downloaded autophagy-related genes from the Human Autophagy Database (HADb). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Screening for autophagy-associated differentially expressed lncRNAs
The autophagy genes were matched with the expression matrix of lncRNAs using Pearson correlation analysis, and lncRNAs with absolute value of correlation coefficient R greater than 0.3 and P value less than 0.001 were selected as autophagy-associated lncRNAs in this study (LncRNA has a large amount of data. In order to filter fewer and more important lncRNA, we set the P value less than 0.001). The grouping was assessed by principal component analysis (PCA).
Construction of autophagy-associated lncRNA prognostic features
One-way Cox regression analysis was performed using the Survival package in R (https://cran.r-project.org/web/packages/survival/index.html) to determine the association of differentially expressed autophagy-associated lncRNAs with patient survival. Autophagy-associated lncRNAs with P<0.05 in the results of one-way Cox regression analysis were subjected to multifactorial Cox regression analysis. Based on the selected lncRNA gene expression multiplied by the sum of the multifactorial Cox regression coefficients, the risk score (risk score) was calculated for each patient, and BRCA patients were divided into low- and high-risk groups according to the median risk value.
Survival analysis
Univariate Cox regression analysis and Kaplan-Meier (KM) method were used to assess the prognostic role of autophagy-associated lncRNA. Survival curves were plotted using the KM method, and log-rank P<0.05 was considered statistically significant.
Correlation analysis of risk scores with immune cells
The level of immune infiltration in BRCA was calculated using the single-sample gene set enrichment analysis (ssGSEA) algorithm, while the correlation between risk score and immune cells for prognostic features was analyzed in the TCGA-BRCA cohort, and finally, the prognosis of immune cell content and risk score was assessed.
Statistical analysis
We used KM plots to assess differential survival between groups, and log-rank P<0.05 was considered statistically significant. Univariate and multivariate (stepwise) Cox regressions were used to construct prognostic characteristics. Wilcoxon test and Spearman’s correlation were used for analysis of variance and correlation, respectively. A P value <0.05 was considered statistically significant.
Results
Results of ARLPs
We extracted 13,413 lncRNAs from the TCGA-BRCA cohort and downloaded 257 autophagy-associated genes HADb. After performing Pierce correlation analysis, we obtained 1,160 autophagy-associated lncRNAs for subsequent analysis.
Results of prognostic characterization of ARLPs
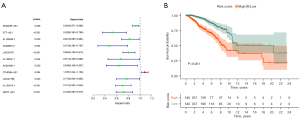
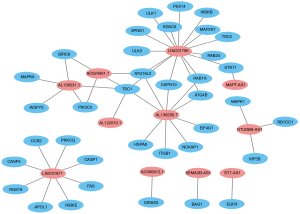
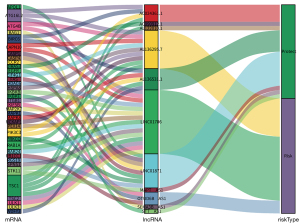
Univariate Cox regression results showed that 42 ARLPs were significantly correlated with OS of BRCA patients. Further multifactorial analysis showed that a total of 11 lncRNAs, including SEMA3B-AS1, ST7-AS1, AL136295.7, AC090912.1, LINC01871, AL136531.1, AC024361.1, OTUD6B-AS1, LINC01786, AL122010.1, and MAPT-AS1, were prognostic independent influencers of BRCA (Figure 1A). Patients were divided into a high-risk group (545 patients) and a low-risk group (545 patients) according to median risk score, and survival analysis showed that patients with low-risk score had a worse prognosis than the high-risk group (P<0.001) (Figure 1B). Subsequently, we constructed a co-expression network of autophagy-related genes by Cytoscape (https://cytoscape.org/) on 11 lncRNAs of the model, which further confirmed that these 11 lncRNAs had significant co-expression relationships (Figure 2). To further understand the relationship between autophagy genes, lncRNAs and risk factors, we performed a clustering analysis. The results showed that LINC01786 and AL136295.7 were the major risk factors, while LINC01871 and AL136531.1 were the major protective factors (Figure 3).
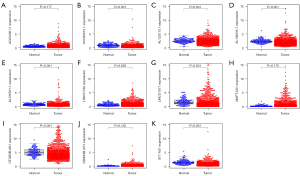
Expression and survival analysis of ARLPs
We analyzed the expression of 11 ARLPs in tumor tissues and normal tissues and showed that AC090912.1, AL122010.1, AL136295.7, AL136531.1, LINC01871, SEMA3B-AS1, and MAPT-AS1 were significantly highly expressed in tumor tissues (P<0.001) (Figure 4). Survival analysis showed that SEMA3B-AS1, ST7-AS1, AL136295.7, AC090912.1, LINC01871, AL136531.1, AC024361.1, LINC01786, AL122010.1, and MAPT-AS1 had a better prognosis in the high-risk score, while OTUD6B-AS1 had a worse prognosis in the high-risk score (Figure 5). This also suggested that OTUD6B-AS1 may be the risk factor while the remaining 10 lncRNAs such as SEMA3B-AS1, ST7-AS1, and AL136295.7 are protective factors.
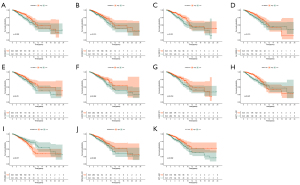
Relationship between risk scores and clinical characteristics
Correlation analysis between risk score and clinical characteristics, univariate analysis showed that risk score was significantly associated with age, clinical stage, tumor size, lymph node metastasis, and distant metastasis (P<0.001). Multifactorial analysis showed that age (P<0.001) was an independent factor affecting risk score (Figure 6A,6B).

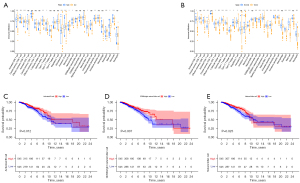
Relationship between risk score and immune cells
We downloaded the results from TCGA database containing various immune cells in BRCA and analyzed the correlation between risk score and immune cells. Grouped according to high- and low-risk scores, the results showed that central memory CD8 T cells, T follicular helper cells, and memory B cell infiltration levels were significantly correlated with high risk scores, and activated CD8 T cells, effector memory CD8 T cells, and type 1 T helper cell, activated B cell, immature B cell, natural killer (NK) T cell, eosinophil, mast cell, and monocyte were significantly associated with low risk scores. The infiltration levels of Activated CD4 T cell, Activated CD8 T cell and Effector memory CD4 T cell were higher in BRCA tissues than in normal tissues. (Figure 7A,7B). Survival analysis showed that activated B cell, CD56 bright NK cell, and NK cell had a better prognosis in the high risk score (Figure 7C-7E).
Discussion
Autophagy can both promote and inhibit tumorigenesis and progression, playing a double-edged role in tumors, as well as those of BRCA. Autophagy provides BRCA cell viability to induce recurrence of metastatic BRCA; whereas, autophagy can also inhibit BRCA metastasis by suppressing cell proliferation (16,17). Several studies have reported that the prognosis of BRCA can be assessed and has some diagnostic value by constructing models for coding genes and non-coding RNAs (18,19). The analyses in these studies were mainly conducted by quantifying the expression levels of transcripts. In our study, lncRNA pairs were generated using autophagy-associated genes and TCGA-BRCA cohort pairing, which is more reliable compared to other studies.
We obtained transcriptomic and clinical data of BRCA patients from TCGA, autophagy genes from HADb database, and extracted autophagy and lncRNA for associated risks. First, differential expression of ARLP was determined and prognostic models were constructed. Immune cell abundance data from the TCGA-BRCA cohort were downloaded from the Tumor Immunity Evaluation Resources (TIMER) database to analyze the relationship between risk scores for ARLP features and immune cell infiltration and prognosis. In our study, 11 autophagy-associated lncRNAs were identified as independent predictors of prognosis in BRCA, and risk models were constructed. Among these 11 autophagy-associated lncRNAs, SEMA3B-AS1, LINC01871, and AL122010.1 have been identified as both BRCA stem cell-associated lncRNAs, which are associated with the prognosis of BRCA patients (20). In addition, studies have shown that SEMA3B-AS1 is involved in the regulation of progression in gastric, hepatocellular, and esophageal cancers (21-23). Studies have shown that the role of ST7-AS1 in tumors is mainly to promote tumor progression. The lncRNA ST7-AS1 promotes malignancy in lung adenocarcinoma by regulating the miR-181b-5p/KPNA4 axis (24). The lncRNA ST7-AS1 promotes TRPM7 expression by upregulating microRNA-543 thereby promoting cervical cancer progression (25). In addition, lncRNA ST7-AS1 expression was found to be downregulated in BRCA, involved in cell cycle, DNA repair, and immune cell infiltration in the BRCA immune microenvironment, and its low expression suggested poor prognosis (26). Studies have shown that OTUD6B-AS1 inhibits thyroid cancer viability, migration, and invasion by targeting miR-183-5p and miR-21 (27). Interestingly, OTUD6B-AS1 regulates the GSKIP/Wnt/β-catenin signaling pathway by segregating miR-664b-3p, thereby promoting cell proliferation and invasion of hepatocellular carcinoma cells (28). As a side note, autophagy-related lncRNAs may play opposite roles in tumors. In addition to this, the inhibitory lncRNA MAPT-AS1 suppresses proliferation and migration and sensitizes cancer cells to paclitaxel by regulating MAPT expression in estrogen receptor (ER)-negative BRCA (29). Other autophagy-associated lncRNAs have not been investigated regarding their biological functions in cancer. Analysis of the correlation of 11 autophagy-associated lncRNAs with clinical characteristics showed significant correlations with age, clinical stage, tumor size, and lymph node status, suggesting that autophagy overarchingly promotes the development and progression of BRCA. Further multifactorial analysis revealed that age was an independent risk factor, with autophagy levels accumulating with age and finally promoting BRCA, as well as explaining the reason why patients with high scoring risk have a worse prognosis.
In the present study, we also focused on the relationship between autophagy-related lncRNA models and immune cell infiltration. The results showed that most of the immune cells were associated with low risk scores, and only some of them were associated with high risk scores. In this study, we focused on the role of T-lymphocyte subpopulations in BRCA. The results showed that most T lymphocytes were concentrated in the low-risk group, suggesting that T lymphocytes may be suppressors of BRCA. For example, CD4+ Th1 secretes interleukins, interferons, and tumor necrosis factor (TNF), which activate effector cells to play an immunoreactive role against BRCA, and NK cells play an oncogenic function during tumor immunosurveillance (30,31). These findings suggest that the role of T lymphocytes may be tuned to different tumor microenvironments. This study was somewhat flawed by the fact that no external data validation was performed, and also by the limitations of the database analysis itself.
Conclusions
In conclusion, this study finally identified 11 autophagy-associated lncRNAs as potential tools for prognosis of BRCA patients and constructed a new model of BRCA prognosis to help clinicians in their assessment of patient prognosis based on risk scores.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6251/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6251/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rodrigues-Ferreira S, Molina A, Nahmias C. Microtubule-associated tumor suppressors as prognostic biomarkers in breast cancer. Breast Cancer Res Treat 2020;179:267-73. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Rossi L, Mazzara C, Pagani O. Diagnosis and Treatment of Breast Cancer in Young Women. Curr Treat Options Oncol 2019;20:86. [Crossref] [PubMed]
- Harbeck N, Gnant M. Breast cancer. Lancet 2017;389:1134-50. [Crossref] [PubMed]
- Liang Y, Zhang H, Song X, et al. Metastatic heterogeneity of breast cancer: Molecular mechanism and potential therapeutic targets. Semin Cancer Biol 2020;60:14-27. [Crossref] [PubMed]
- Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013;193:651-69. [Crossref] [PubMed]
- Pang B, Wang Q, Ning S, et al. Landscape of tumor suppressor long noncoding RNAs in breast cancer. J Exp Clin Cancer Res 2019;38:79. [Crossref] [PubMed]
- Hua Q, Mi B, Xu F, et al. Hypoxia-induced lncRNA-AC020978 promotes proliferation and glycolytic metabolism of non-small cell lung cancer by regulating PKM2/HIF-1α axis. Theranostics 2020;10:4762-78. [Crossref] [PubMed]
- Yan X, Hu Z, Feng Y, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell 2015;28:529-40. [Crossref] [PubMed]
- Noda NN, Inagaki F. Mechanisms of Autophagy. Annu Rev Biophys 2015;44:101-22. [Crossref] [PubMed]
- White E. The role for autophagy in cancer. J Clin Invest 2015;125:42-6. [Crossref] [PubMed]
- Sun T. Long noncoding RNAs act as regulators of autophagy in cancer. Pharmacol Res 2018;129:151-5. [Crossref] [PubMed]
- Amelio I, Bernassola F, Candi E. Emerging roles of long non-coding RNAs in breast cancer biology and management. Semin Cancer Biol 2021;72:36-45. [Crossref] [PubMed]
- Soysal SD, Tzankov A, Muenst SE. Role of the Tumor Microenvironment in Breast Cancer. Pathobiology 2015;82:142-52. [Crossref] [PubMed]
- Vera-Ramirez L, Vodnala SK, Nini R, et al. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun 2018;9:1944. [Crossref] [PubMed]
- Chang CH, Bijian K, Wernic D, et al. A novel orally available seleno-purine molecule suppresses triple-negative breast cancer cell proliferation and progression to metastasis by inducing cytostatic autophagy. Autophagy 2019;15:1376-90. [Crossref] [PubMed]
- Chang JT, Wang F, Chapin W, et al. Identification of MicroRNAs as Breast Cancer Prognosis Markers through the Cancer Genome Atlas. PLoS One 2016;11:e0168284. [Crossref] [PubMed]
- Xiao B, Zhang W, Chen L, et al. Analysis of the miRNA-mRNA-lncRNA network in human estrogen receptor-positive and estrogen receptor-negative breast cancer based on TCGA data. Gene 2018;658:28-35. [Crossref] [PubMed]
- Li X, Li Y, Yu X, et al. Identification and validation of stemness-related lncRNA prognostic signature for breast cancer. J Transl Med 2020;18:331. [Crossref] [PubMed]
- Dong Z, Liang X, Wu X, et al. Promoter hypermethylation-mediated downregulation of tumor suppressor gene SEMA3B and lncRNA SEMA3B-AS1 correlates with progression and prognosis of esophageal squamous cell carcinoma. Clin Exp Metastasis 2019;36:225-41. [Crossref] [PubMed]
- Guo W, Liang X, Liu L, et al. MiR-6872 host gene SEMA3B and its antisense lncRNA SEMA3B-AS1 function synergistically to suppress gastric cardia adenocarcinoma progression. Gastric Cancer 2019;22:705-22. [Crossref] [PubMed]
- Zhong Y, Li Y, Song T, et al. MiR-718 mediates the indirect interaction between lncRNA SEMA3B-AS1 and PTEN to regulate the proliferation of hepatocellular carcinoma cells. Physiol Genomics 2019;51:500-5. [Crossref] [PubMed]
- Hu RH, Zhang ZT, Wei HX, et al. LncRNA ST7-AS1, by regulating miR-181b-5p/KPNA4 axis, promotes the malignancy of lung adenocarcinoma. Cancer Cell Int 2020;20:568. [Crossref] [PubMed]
- Qi H, Lu L, Wang L. Long Noncoding RNA ST7-AS1 Upregulates TRPM7 Expression by Sponging microRNA-543 to Promote Cervical Cancer Progression. Onco Targets Ther 2020;13:7257-69. [Crossref] [PubMed]
- Zhang Z, Zhang H, Li D, et al. LncRNA ST7-AS1 is a Potential Novel Biomarker and Correlated With Immune Infiltrates for Breast Cancer. Front Mol Biosci 2021;8:604261. [Crossref] [PubMed]
- Wang Z, Xia F, Feng T, et al. OTUD6B-AS1 Inhibits Viability, Migration, and Invasion of Thyroid Carcinoma by Targeting miR-183-5p and miR-21. Front Endocrinol (Lausanne) 2020;11:136. [Crossref] [PubMed]
- Kong S, Xue H, Li Y, et al. The long noncoding RNA OTUD6B-AS1 enhances cell proliferation and the invasion of hepatocellular carcinoma cells through modulating GSKIP/Wnt/β-catenin signalling via the sequestration of miR-664b-3p. Exp Cell Res 2020;395:112180. [Crossref] [PubMed]
- Pan Y, Pan Y, Cheng Y, et al. Knockdown of LncRNA MAPT-AS1 inhibites proliferation and migration and sensitizes cancer cells to paclitaxel by regulating MAPT expression in ER-negative breast cancers. Cell Biosci 2018;8:7. [Crossref] [PubMed]
- Maxwell AJ, Clements K, Hilton B, et al. Risk factors for the development of invasive cancer in unresected ductal carcinoma in situ. Eur J Surg Oncol 2018;44:429-35. [Crossref] [PubMed]
- Edechi CA, Ikeogu N, Uzonna JE, et al. Regulation of Immunity in Breast Cancer. Cancers (Basel) 2019;11:1080. [Crossref] [PubMed]
(English Language Editor: J. Jones)


