
Peritoneal dialysis effluent-derived exosomal miR-432-5p: an assessment tool for peritoneal dialysis efficacy
Introduction
Peritoneal dialysis (PD) is an effective alternative renal replacement therapy for patients with end-stage renal disease. Patients undergoing PD vary significantly in terms of their peritoneal solute transport rate (PSTR), which is evaluated by the peritoneal equilibration test (PET) (1). Higher PSTRs are associated with declines in ultrafiltration (UF) volume and dialytic sodium removal (DSR), resulting in poor patient treatment. Adequate fluid clearance requires the removal of water coupled with sodium and is crucial for a high-quality PD (2). Overhydration and hypertension are commonly reported in PD patients (3). Both optimized UF and DSR are associated with a reduction in patient mortality (4). Sun et al. (5) demonstrated that excessive salt intake under uremic conditions induced functional and structural changes in the peritoneal membrane, leading to higher baseline PSTRs in subtotal nephrectomized mice. Our preliminary data showed that patients with different PSTRs have different dialytic sodium concentrations, but the relationship and molecular mechanisms remain unexplored.
Exosomes are small membrane-bound extracellular vesicles (EVs) with diameters of 30–150 nm. They are secreted by different types of cells, found in many biological fluids (6), and are composed of a lipid bilayer and abundant cellular cargos, such as nucleic acids [DNA, messenger RNA (mRNA), microRNA (miRNA), and other non-coding RNA (ncRNA)], proteins, and bioactive lipids that can be transferred between cells (7). Since the contents and functions of exosomes derived from different cell types are distinct, exosomes can act as biomarkers of various diseases, such as acute kidney injury (AKI) and renal cell carcinoma (8).
The miRNA are a class of small, endogenous, single-stranded ncRNAs which regulate gene expression at the post-transcriptional level (9). Actively secreted miRNAs are packaged into exosomes and involved in cell-cell communication (10).
Peritoneal dialysis effluent (PDE) is easily collected from PD patients for a variety of analyses. Although previous studies have illustrated that proteins in exosomes derived from PDE are related to PSTR and PD efficiency (11,12), there has been no determination that exosomal miRNA in PDE (PDE-exo-miRNA) has any effect on, or reflects, PSTR or PD efficiency. It is also unknown whether PDE-exo-miRNA is involved in UF or DSR.
The epithelial sodium channel (ENaC) is a non-voltage-gated sodium ion channel in the apical membrane of polarized epithelial cells (13). It plays an important part in maintaining the dynamic balance of sodium and water by regulating blood volume and blood pressure, renal sodium reabsorption, and the airway surface fluid clearance of lung epithelial cells (14-17). The expression and function of ENaC can be influenced by multiple factors, such as serum glucocorticoid-induced kinase 1, aldosterone, drugs, and miRNA (16-19). To date, the relationship between PDE-exo-miRNA and ENaC has not been explored.
In this study, we used miRNA array to investigate the miRNA profiles of PDE-exo in patients with different PSTRs, as well as the potential mechanism of DSR. We selected the significantly differential miR-432-5p, and its target gene ENaC α subunit (α-ENaC), according to bioinformatics analysis. We measured the expression of the significantly differential miR-432-5p in 40 patients and determined its correlation to PSTR, 4-hour (4 h) UF, and DSR. We also verified α-ENaC as a potential target gene of miR-432-5p. Consequently, we propose that miR-432-5p may target α-ENaC, thus affecting the processes of UF and DSR in PD.
We present the following article in accordance with the Materials Design Analysis Reporting (MDAR) checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-3957/rc).
Methods
Sample collection
The PET is a key tool to assess a patient’s PSTR, which measures the dialysate/plasma ratio of creatinine after a standardized 4 h dwell (4 h D/PCr) of 3.86% glucose dialysate (20). In this study, patients underwent PET at 6-month intervals and were classified into 4 transport characteristics according to 4 h D/PCr: high (H; 0.82–1.03), high average (HA; 0.65–0.81), low average (LA; 0.50–0.65), and low (L; 0.34–0.49). The PSTR was considered stable if a patient returned the same H/HA or LA/L result 3 times consecutively. Our cross-sectional study aimed to investigate the relationship between PDE-exo-miRNA and UF volumes and PSTRs of PD patients. We enrolled 50 patients who received regular continuous ambulatory peritoneal dialysis (CAPD) at Shanghai Ninth People’s Hospital between January 2000 and July 2020. Patients (aged 18–80 years) with stable PSTRs and nocturnal dwell prescriptions were included in our study. None of the participants had peritonitis 3 months prior to, or during, the study. The exclusion criteria were malignancies, abdominal operation histories, autoimmune diseases, hepatitis, tuberculosis, pulmonary infections, or systematic inflammation. The patients selected for the study were divided into 2 groups, H (H/HA) and L (LA/L), according to the results of their PETs.
We explained the aim of our study to the patients included in this study and guaranteed their privacy. All individual participants signed and provided informed consent for the use of their PDE and clinical data for research purposes. The study was conducted according to the tenets of the Declaration of Helsinki (as revised in 2013) and approved by the Ethics Committee of Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine (SH9H-2020-T23-1).
The nocturnal PDE was collected before PET was performed. We weighed and recorded the drained volumes after 4 h PET and the 24 h total UF volumes before PET. The sodium concentrations in PDE were also measured. The DSR was calculated using the following formula:
where Vd denotes the drained volume, Vi the infused volume, and Cd and Ci are the sodium concentrations in Vd and Vi, respectively.
Exosome isolation and characterization
We first collected the overnight PDE and shook it well to ensure the uniformity of its components. As EVs in the peritoneal cavity can be diluted in the PDE after 2 L of peritoneal dialysate infusion, we used a Stirred Ultrafiltration Cell with a 10 kDa NMWL, Ultracel regenerated cellulose membrane (MilliporeSigma, Billerica, MA, USA) to concentrate 500 mL of the PDE. Since the membrane was 10 kDa NMWL, the EVs were expected to remain in the liquid above the membrane, while most of the water and other small molecules would pass through the filter. The concentrate was centrifuged at 2,500 g for 15 min at 4 ℃ to remove cellular debris. After centrifugation at 18,000 g for 30 min at 4 ℃, the supernatants were filtered through a 0.22-µm filter to remove large substances. The samples were centrifuged at 120,000 g for 2 h at 4 ℃ to obtain exosomes using an Optima XE-90 ultracentrifuge (Beckman Coulter, Inc., Indianapolis, IN, USA). The pellets were then washed again with a phosphate buffer solution (PBS) at 120,000 g for 2 h. The exosomes were stored at −80 ℃ for further experiments.
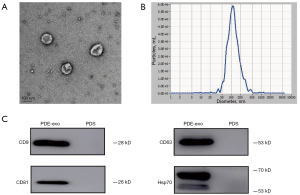
To identify and observe the morphology of the exosomes, we used a Tecnai F20 transmission electron microscopy (TEM; FEI Company, Hillsboro, OR, USA). The particle sizes were evaluated using ZetaView Nanoparticle Tracking Analysis (NTA; Particle Metrix GmbH, Inning am Ammersee, Germany). Western blot was conducted to detect the specific markers that identify exosomes.
RNA extraction and microarray
We extracted RNA was from PDE-exos using an miRNeasy Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. To qualify all the extracted RNA, we performed quality control of the miRNA using an Agilent 4200 TapeStation (Agilent Technologies, Santa Clara, CA, USA) and measured the A260/A280 using NanoDrop Microvolume Spectrophotometers. We added 3.5 µL cel-miR-39 to the denatured exosomal mixture according to the manufacturer’s protocols (exoRNeasy Serum/Plasma Maxi Kits; QIAGEN). Exosomal miRNAs from 10 different patients (group H, n=5; group L, n=5) were purified and quality assessed. The differentially expressed miRNAs were screened and analyzed for more details using a GeneChip miRNA 4.0 Array (Affymetrix Inc., Sunnyvale, CA, USA).
Bioinformatics analysis
For bioinformatics analysis, we first used the miTarBase 7.0 (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) database to obtain the target genes of miRNAs. We then used Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses on the Database for Annotation, Visualization, and Integrated Discovery (DAVID) 6.8 platform (https://david.ncifcrf.gov/) to determine the possible biological functions and signaling pathways. The GO analysis was classified into 3 categories, biological process (BP; red), molecular function (MF; blue), and cellular component (CC; green). The threshold of enrichment analysis was set as P<0.05. Lastly, Targetscan Human 7.2 (http://www.targetscan.org/vert_72/), microrna.org (http://www.microrna.org/microrna/home.do), miRDB (http://mirdb.org/), and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) were used to determine the target genes of differentially expressed miRNAs.
Cell culture and transfection
Human MeT-5A mesothelial cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). They were cultured in an M199 medium (Hyclone Laboratories, Logan, UT, USA) with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin (Biological Industries, Kibbutz Beit HaEmek, Israel) at an atmosphere of 5% CO2 at 37 ℃. The cells were transfected with an miR-432-5p mimic, inhibitor, and respective negative control (NC) using a riboFECT CP Transfection Kit (RiboBio, Guangzhou, China). The dose and procedure were administered according to the manufacturer’s protocols. Each experiment was performed with 3 independent replicates.
Reverse transcription and quantitative real-time polymerase chain reaction
The miRNA extracted from PDE was polyadenylated and reverse transcribed into complementary DNA (cDNA) with the miDETECT A Track miRNA qRT-PCR Starter Kit (RiboBio). The miRNA extracted from cells was reverse transcribed using a stem-loop primer, and mRNA was reverse transcribed using a PrimeScript RT Reagent Kit (Takara Bio Inc., Shiga, Japan). The products of reverse transcription polymerase chain reaction (RT-PCR) were used to perform agarose gel electrophoresis. We performed SYBR Green PCR amplification on a real‐time PCR system (Thermo Fisher Scientific, Waltham, MA, USA) using a TB Green Premix Ex Taq kit (Takara Bio Inc.). The relative expression level of exosomal miRNA was normalized to cel-miR-39. After transfection, cell miRNA was normalized to U6 (common control gene), and the cell mRNA was normalized to housekeeping gene glyceraldehyde 3-phospahte dehydrogenase (GAPDH). We calculated the relative expression levels using the equation:
Western blot analysis
Samples of exosomes or MeT-5A cells protein lysate were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on 10% gels and transferred onto a polyvinylidene difluoride (PVDF) membrane. Blots were blocked with 5% nonfat milk and then incubated overnight at 4 ℃ with the primary antibodies CD9, CD63, CD81, Hsp70 (1:1,000; SBI Medical, Fort Lauderdale, FL, USA), α-ENaC (1:500; Alomone Labs, Jerusalem, Israel), or GAPDH (1:5,000; Proteintech Group Inc., Rosemont, IL, USA). They were then blotted at room temperature (RT) for 1 h with horse radish peroxidase (HRP)-conjugated Affinipure goat anti-rabbit immunoglobulin G (IgG) secondary antibody (1:5,000; Proteintech Group Inc.). The blots were visualized using an enhanced chemiluminescence detection system.
Immunohistochemistry (IHC)
Specimens of human peritoneum were obtained from patients who underwent elective abdominal surgery. After antigen retrieval, 3% hydrogen peroxide was added to block endogenous peroxidase activity. The slides were washed with PBS and incubated with the α-ENaC antibody at 4 ℃ overnight. Then, PBS was used in place of the primary antibody as NC. The slides were incubated with a secondary antibody at 37 ℃ for 30 min the next day. Immunohistochemical Strept Avidin-Biotin Complex (Solarbio, Beijing, China) was used according to the manufacturer’s protocols. After diaminobenzidine staining and hematoxylin counterstaining, the slides were observed under a microscope.
Dual luciferase reporter assay
To verify that SCNN1A is the target gene of miR-432-5p, HEK-293T cells were co-transfected with SCNN1A-wild-type (WT), mutant (MUT), or NC PmirGLO vectors, and miR-432-5p mimic or corresponding NC, using a Lipofectamine 3000 Reagent (Thermo Fisher Scientific) based on the manufacturer’s instructions. After 48 h transfection, a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) was utilized to measure the luciferase activities. The ratio of renilla luciferase activity to firefly luciferase activity was calculated. Each treatment was repeated 3 times.
Cell Counting Kit-8 (CCK-8) assay
The MeT-5A cells were seeded into 96-well plates and transfected with NC, mimic, or inhibitor for 48 h. Each group of cells were then incubated with 10 µL CCK-8 reagents (Biosharp, Shanghai, China) for 2 h at 37 ℃. The viability of cells was measured by absorbance values at 450 nm.
Fluorescent labeling of exosomes
To observe how PDE-exo-miRNA influences MeT-5A cells, we labeled the PDE-exo with a PKH67 Green Fluorescent Cell Linker Kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions. The MeT-5A cells were incubated with the labeled exosomes or control solution overnight. The cell slides were washed with PBS and fixed with 4% formaldehyde solution at RT for 10 min. The cytoskeleton was then stained with TRITC-phalloidin (Solarbio), and the nuclei of the MeT-5A cells were stained by 4',6-diamidino-2-phenylindole (DAPI) (Beyotime Biotechnology, Shanghai, China). We observed the uptake and internalization of the labeled exosomes with the use of a fluorescence microscope.
Statistical analysis
All statistical analyses and graph generation were performed using SPSS Statistics 23 (IBM Corp., Armonk, NY, USA) and Graphpad Prism 8.0.1 (GraphPad Software, San Diego, CA, USA). Measurement data in accordance with Gaussian distribution were represented as mean ± SD, and an unpaired, 2-tailed Student’s t-test was used to compare the 2 groups. Non-Gaussian distribution data are represented as M (1/4, 3/4), and a Mann-Whitney U test was used to compare the 2 groups. Enumeration data were analyzed by Fisher’s exact test or chi-square test. One-way analysis of variance (ANOVA) was performed to compare multiple groups with different treatments. Spearman correlation was used for correlation analysis. A P value <0.05 was considered statistically significant. The enriched GO terms and KEGG pathways were visualized using RStudio software (RStudio, Boston, MA, USA). The Bingo plugin in the Cytoscape software (https://cytoscape.org/) was applied to describe the GO analysis network.
Results
PDE-exo identification
To identify whether the EVs isolated from concentrated overnight PDE by ultracentrifugation were exosomes, we first observed them under TEM and observed the characteristic cup-shaped vesicles with a bilayer structure (Figure 1A). The results of NTA showed that the peak diameter of purified particles was approximately 100 nm (Figure 1B). We found that the specific surface markers of exosomes, such as CD9, CD63, CD81, and Hsp70 were positive (Figure 1C). Taken together, our findings suggested that the PDE-derived EVs collected in our study were exosomes.
Different PDE-exo miRNA profiles
The demographic and clinical characteristics of the 10 participants used for miRNA array are summarized in Table 1. Apart from 4 h D/PCr, there were no significant differences in age, gender, dialysis time, estimated glomerular filtration rate (eGFR), and Kt/V between the 2 groups.
Table 1
| Clinical characteristics | H group (n=5) | L group (n=5) | Test value | P value |
|---|---|---|---|---|
| Gender (male/female) | 2/3 | 1/4 | –a | >0.999 |
| Age (years) | 63.40±15.96 | 57.40±19.37 | 0.535b | 0.608 |
| Dialysis time (months) | 38.20±12.58 | 39.80±15.27 | 0.181b | 0.861 |
| 4 h D/PCr | 0.77±0.04 | 0.58±0.07 | 5.364b | 0.0007*** |
| eGFR (mL/min/1.73 m2) | 3.57±3.42 | 2.64±3.58 | 0.424b | 0.683 |
| Kt/V (mL/min) | 2.00±0.47 | 2.17±0.89 | 0.386b | 0.710 |
Values are means ± SD. a, Fisher’s exact test; b, t-test; ***, P<0.001. eGFR, estimated glomerular filtration rate; Kt/V, urea clearance index; SD, standard deviation.
The miRNA expression profiles of PDE-exo from patients with different PSTRs was characterized by miRNA array. We used the limma package in R software to screen different genes before evaluating the fold change (FC) and significance level of the differences between group H and L. The miRNA microarray found 590 miRNAs and 8 miRNAs were significantly different (P<0.05) in group H when compared to group L. Among them, the 5 miRNAs with FC >1.7 were upregulated ones, and 3 miRNAs with FC <0.63 were downregulated ones. Cluster analysis was shown based on the signal values of each differentially expressed miRNA in the samples (Figure 2A,2B). The top 10 GO terms in each GO category and the significant pathway terms are listed in Figure 2C,2D. The GO analysis of upregulated miRNAs showed that GO terms relevant to cellular complexes and protein binding were predominantly enriched in the CC and MF. Interestingly, the second and third items in the BP part were related to sodium transport. Therefore, we focused on the upregulated miRNAs to predict their target genes. The GO and KEGG analysis of downregulated miRNAs are provided in Figure S1.
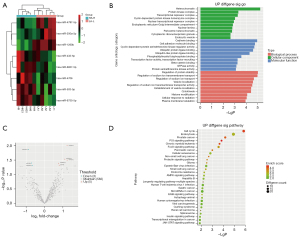
Bioinformatic prediction of upregulated miRNA
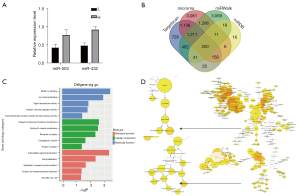
Among the 5 upregulated miRNAs, miR-503-5p and miR-432-5p were the most enriched miRNAs according to the volcano plot (Figure 2A). The results showed that when compared with group L, a 1.72-fold higher level of miR-503-5p expression, and a 1.91-fold higher level of miR-432-5p expression, were observed in group H (Figure 3A). For this reason, we selected miR-432-5p for further analysis. A Venn diagram was used to identify the overlapping target genes of miR-432-5p predicted by Targetscan Human, mirnaorg, miRDB, and miRWalk (Figure 3B). The GO and KEGG pathway enrichment analyses were implemented with DAVID for genes present in 3 or 4 concurrent databases. The GO analysis revealed that the candidate targets of miR-432-5p were involved in metal ion binding, sodium channel regulator activity, and sodium ion transmembrane transport (Figure 3C). Furthermore, an enriched GO terms network was established using the Bingo plugin in Cytoscape software. Ion transport and ion channel activity were also illustrated by this network (Figure 3D). The whole image of the Bingo network is provided in the supplementary material (Figure S2). Based on the miRNA profiling data and bioinformatic predictions, we determined to investigate the role of miR-432-5p in sodium transport.
Validation of PDE-exo-miR-432-5p by quantitative RT-PCR (qRT-PCR)
To confirm miR-432-5p was differentially expressed in the H and L groups, qRT-PCR analysis was performed in the validation cohorts (20 patients in each group). There were no significant differences in gender, age, dialysis time, eGFR, Kt/V, hypertension, and diabetes between group H and L. However, a significant difference was found between the 2 groups in terms of the 4 h D/PCr (Table 2). The results of qRT-PCR analysis showed that there was a 1.86-fold higher level of relative PDE-exo-miR-432-5p expression (P=0.002) in group H compared with group L (Figure 4A).
Table 2
| Clinical characteristics | H group (n=20) | L group (n=20) | Test value | P value |
|---|---|---|---|---|
| Gender (male/female) | 10/10 | 8/12 | 0.404a | 0.525 |
| Age (years) | 59.10±15.33 | 58.20±13.47 | 0.197b | 0.845 |
| Dialysis time (months) | 45.10±21.37 | 54.55±23.86 | 1.319b | 0.195 |
| 4 h D/PCr | 0.76±0.06 | 0.58±0.04 | 11.390b | <0.0001**** |
| eGFR (mL/min/1.73 m2) | 2.44 (0, 4.25) | 0 (0, 3.84) | 173.50c | 0.446 |
| Kt/V (mL/min) | 2.00 (1.69, 2.41) | 1.80 (1.60, 1.94) | 0.386c | 0.710 |
| Hypertension (yes/no) | 18/2 | 13/7 | –d | 0.127 |
| Diabetes (yes/no) | 8/12 | 6/14 | 0.300ª | 0.584 |
Values are means ± SD or M (1/4, 3/4). a, chi-square test; b, t-test; c, Mann-Whitney U test; d, Fisher’s exact test; ****, P<0.0001. eGFR, estimated glomerular filtration rate; Kt/V, urea clearance index; SD, standard deviation.
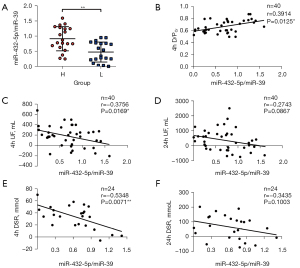
PDE-exo-miR-432-5p is associated with PET, UF volume, and DSR
Since miR-432-5p was increased in group H, we hypothesized that miR-432-5p might correlate with 4 h D/PCr and UF volume. Spearman correlation analysis indicated that a relative miR-432-5p expression level was positively correlated with 4 h D/PCr (r=0.391; P<0.05) (Figure 4B). We observed a negative correlation between PDE-exo-miR-432-5p and 4 h UF (r=−0.376; P<0.05). However, PDE-exo-miR-432-5p was not correlated with 24 h UF (Figure 4C,4D). We also observed a negative correlation between PDE-exo-miR-432-5p and 4 h DSR (r=−0.535; P<0.01), but there was no relation between PDE-exo-miR-432-5p and 24 h DSR (Figure 4E,4F).
α-ENaC is expressed on both human peritoneum and MeT-5A cells
Bioinformatic prediction indicated that 12 target genes of miR-432-5p were associated with sodium ion transmembrane transport (Table 3). However, only SCNN1A has a broad expression in the epithelial tissues of various organs and encodes the α-subunit of ENaC, which is an indispensable component of nonselective channels contributing to alveolar fluid clearance (21).
Table 3
| Sodium ion transmembrane transport (GO:0035725) | Genes |
|---|---|
| Voltage-gated sodium channel (SCN) | SCN4A, SCN5A, SCN3B |
| Epithelial sodium channel (ENaC) | SCNN1A, SCNN1G |
| Solute carrier family (SLC) | SLC4A4, SLC12A3, SLC24A1, SLC24A2, SLC34A1 |
| Others | ASIC2, HCN4 |
GO, Gene Ontology.
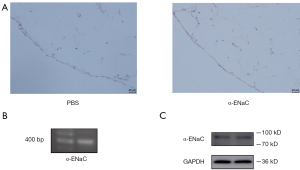
The expression of α-ENaC on human peritoneum was determined by IHC. It was revealed that α-ENaC was primarily localized to monolayer mesothelial cells of peritoneum (Figure 5A). The result of RT-PCR showed that SCNN1A (406 bp), which encodes α-ENaC, was present in Met-5A cells (Figure 5B). A band at 90 kDa corresponding to α-ENaC was revealed in Met-5A cells (Figure 5C).
SCNN1A is a direct target gene of miR-432-5p
Targetscan predicted a complementary sequence between miR-432-5p and SCNN1A (Figure 6A). Therefore, to verify whether SCNN1A is a target gene of miR-432-5p, MeT-5A cells were transfected with miR-432-5p mimic or inhibitor. When a mimic was transfected, the expression level of miR-432-5p relative to U6 significantly increased, but miR-432-5p expression did not change in either the blank or NC group. The levels of α-ENaC mRNA and protein exhibited a marked decrease in the mimic group compared to the control group (Figure 6B,6C), while α-ENaC mRNA and protein were increased by inhibiting miR-432-5p (Figure 6D,6E). We also found that Met-5A cell viability was not influenced by transfection (Figure 6F).
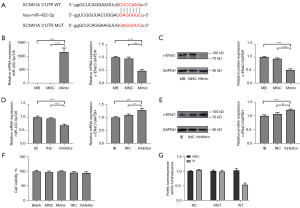
In addition, a dual luciferase reporter assay was applied to elucidate the targeting relationship between miR-432-5p and α-ENaC. The 293T cells were transfected with plasmids and mimic/NC. We found that the relative luciferase activity in the WT + miR-432-5p mimic group was significantly lower than that in any other groups, which confirmed that miR-432-5p directly interacted with the binding sites at the 3'untranslated region (3'-UTR) of SCNN1A mRNA (Figure 6G).
PDE-exo uptake by MeT-5A cells
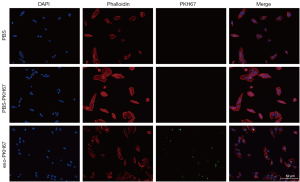
To investigate if PDE-exo-miR-432-5p affected DSR, we labeled the exosomes with PKH67, which is a green-fluorescent dye. We then added them to the cell medium to observe whether the PDE-exo could be taken into cells. We used PKH67-PBS and PBS controls as the reference. After 24 h incubation, the exosomes were incorporated into the MeT-5A cells and distributed in the cytoplasm. However, this did not occur for the control groups, which showed no fluorescence (Figure 7). These observations clarified that PDE-exo can be internalized and is thus a potential regulator of genetic expression in MeT-5A cells.
Discussion
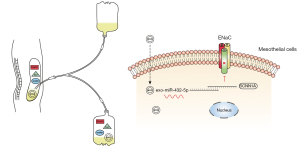
In this study, we isolated EVs from concentrated PDE and found they possessed a size and composition characteristic of exosomes (22). Their morphology and size distribution were also consistent with the findings of other studies (11,12). For this reason, we used the term exosome when describing these vesicles. Examining the exosomal miRNA profiles in patients with different PSTRs yielded evidence that the expression level of PDE-exo-miR-432-5p was higher in group H than that in group L, and this was negatively correlated to 4 h UF and DSR. Furthermore, we verified that SCNN1A is one of the target genes of miR-432-5p. Since PDE-exo can be internalized into mesothelial cells, they can therefore be regarded as carriers of miRNA, which inhibit the expression of α-ENaC (Figure 8).
It is suggested that the criterion for assessing PD adequacy should include not only solute clearance, such as urea or creatine clearance, but also fluid and sodium removal, control of blood pressure, and adequate nutrition (4). Previous trials in PD have demonstrated that sodium and fluid overload result in a higher risk for cardiovascular events and technique failure (23). Patients with higher PSTRs are more prone to sodium and water retention than those with lower PSTRs, thus leading to UF failure and a poor prognosis (2). Although patients can achieve similar Kt/V by adjusting the prescriptions of PD to meet the standard dialysis adequacy, we found that the differences of DSR among patients with different PSTR may affect UF and result in volume overload. In recent years, many long non-coding (lnc)RNAs/miRNAs have been identified as diagnostic or prognostic biomarkers of human diseases. For example, urinary exosomal ncRNAs-miRNA-181a is a diagnostic biomarker for the early detection of chronic kidney disease (CKD). Both HCP5 and lncRNA NOP14-AS1 are potential biomarkers to predict CKD progression. Furthermore, there are various lncRNAs involved in experimental AKI that have been noted to improve renal injury by reducing inflammation, oxidative stress, and cell death, such as LRNA9884, LINC00520, and so on (24). A previous study found that lncRNA 6030408B16RIK silencing can prevent UF failure in PD via miRNA-326-3p-mediated WISP2 down-regulation (25). However, few miRNAs or lncRNAs can predict peritoneal transport characteristics or have been reported as being involved in UF and DSR. To extend the research on this topic, we used an miRNA array to analyze PDE-exo-miRNA profiles from patients with different PSTRs in this study.
Exosomes are well recognized as carriers of miRNA and protein delivery (26,27). Szeto et al. (28) found that a change in PDE-miR-21 and miR-589 levels over 1 year produced a corresponding change in 4 h D/PCr. However, there was no significant correlation between these miRNAs and UF volume. Unlike these previous studies, which extracted free miRNA directly from PDE (28,29), we extracted exo-miRNA, which has a higher stability and resistance to degradation due to the bilayer structure of exosomes. We first examined the differentially expressed PDE-exo-miRNAs in patients from H and L PSTR groups. We found the expression of miR-432-5p showed the most variation when compared to several other differential miRNAs measured. As GO and KEGG pathway analyses suggested that the target genes of miR-432-5p may mediate sodium transport, we enrolled 40 patients to investigate whether miR-432-5p is involved in the regulation of UF and DSR. Our study showed that PDE-exo-miR-432-5p was significantly higher in group H and had a weak positive correlation with 4 h D/PCr. However, PDE-exo-miR-432-5p had a moderate negative correlation with 4 h UF and 4 h DSR, but not with 24 h UF or 24 h DSR. Our results surrounding the correlation between PDE-exo-miR-432-5p and UF or DSR were thus inconsistent at 4 and 24 h dwell times. Because 24 h UF and 24 h DSR were also affected by PD conditions (glucose concentration, dialysis time, dialysis fluid dwell time), urine volume, and dietary sodium intake, we took the 4 h dwell time results as being more valid. It is yet to be determined if PDE-exo-miR-432-5p can be used as a reliable biomarker for measuring PSTR. Large sample clinical trials and more experiments are needed to test the sensitivity and specificity of miR-432-5p.
Previous studies have shown that miR-432-5p plays an important role in the regulation of the development and drug resistance of cancers (30,31); however, the function of miR-432-5p in sodium ion transport remains unclear. To eliminate the limitations of a single database, we used 4 different databases to predict the target genes and investigated whether the target genes of miR-432-5p could influence sodium ion transport in PD. Our analysis indicated that the primary target genes of miR-432-5p included the voltage-gated sodium channel (SCN), ENaC, solute carrier family (SLC), acid sensing ion channel (ASIC), and hyperpolarization activated cyclic nucleotide gated potassium channel 4 (HCN4) (Table 3). The SCN4A, SCN5A, and SCN3B genes are expressed in skeletal and cardiac muscle (32). The SLCs are among the target genes of miR-432-5p and encode different sodium channels or transporters which have restricted expressions in the kidney, rods, and cones of the retina (33-36). The ASIC gene mainly expresses in brain and endometrium (37), whereas HCN4 encoded proteins are necessary for cardiac pacing (38). Since the ENaC gene is the only one broadly expressed in the epithelium over the body, we focused our experiment on whether it was the target gene of miR-432-5p. Although both the α-ENaC gene SCNN1A and the γ-ENaC gene SCNN1G are putative target genes of miR-432-5p, we preferred SCNN1A for the potential it showed for binding better to miR-432-5p (Figure S3). In addition, α subunit is vitally important for ENaC composition. In a previous study, mice deficient in α-ENaC were found to die of respiratory distress more quickly than those lacking the γ subunit, indicating that α-ENaC plays a decisive role in lung fluid clearance (39). On account of these factors, we proposed that α-ENaC may be critical in determining the DSR in PD.
The channels and transporters on mesothelial cells are responsible for regulating the transport of fluids and electrolytes across serous membranes (40). Nie et al. (41) demonstrated that the ENaC channels were expressed in human pleural mesothelial cells. Peritoneal mesothelium is a part of the barriers responsible for solute transport in PD. In the present study, we detected the expression of α-ENaC on human peritoneum biopsies and mesothelial cells. Subsequently, we demonstrated that α-ENaC is the target of miR-432-5p. To investigate how PDE-exo-miR-432-5p participate in DSR, we observed PDE-exo internalization, suggesting that the PDE-exo can transfer information to the mesothelial cells. This may, in turn, inhibit the expression of α-ENaC and decrease DSR (Figure 8).
The rate of technique failure in patients with high PSTR is much higher than that in those with low PSTR. Some studies have shown that increased levels of interleukin-6 (IL-6), vascular endothelial growth factor (VEGF), and matrix metalloproteinase-2 (MMP-2) are indicative of high transport characteristics. In our study, we found that patients have different PDE-exo-miRNA profiles depending on their PSTRs. However, whether PDE-exo-miR-432-5p can be used as a biomarker for measuring PSTR will require large sample clinical trials. In addition, PDE-exo-miR-432-5p may play an important role in DSR by inhibiting α-ENaC, but the sensitivity and specificity of miR-432-5p in predicting sodium and water clearance require further investigation. To this end, animal experiments could be conducted to characterize the role of ENaC in DSR and dissect the underlying molecular pathways. Future studies are also necessary to explore the mechanism of how miR-432-5p is regulated and what signal pathway it uses to effect sodium removal.
Conclusions
In conclusion, our study identified a unique profile for exosomal miRNAs in the PDE of patients with different peritoneal membrane transport characteristics. We found that the relative expression level of PDE-exo-miR-432-5p was higher in the H PSTR group, and that the PDE-exo-miR-432-5p expression level was negatively correlated with 4 h UF and 4 h DSR. We also demonstrated that α-ENaC is a gene for miR-432-5p targeted inhibition. Our findings provide evidence that PDE miR-432-5p could be used as a biomarker to determine the effectiveness of PD and provide a novel prospect for the mechanism of sodium removal during PD.
Acknowledgments
We gratefully thank all the patients for their kind participation in the study and other members of our laboratory. We express special appreciation to Professor Delon Barfuss and Professor Yanhua Wang for polishing our manuscript.
Funding: This work was supported by the Clinical Research Program (JYLJ2018011) funding of Ninth People’s Hospital affiliated to Shanghai Jiao Tong University School of Medicine, National Natural Science Foundation of China (8217030178) and Shanghai Health Commission Clinical Research Project (202140430). The funders were not involved in the study design, data collection, data analysis, interpretation, or writing of the report.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-3957/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-3957/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-3957/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine (SH9H-2020-T23-1) and informed consent was provided by all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Asano M, Ishii T, Hirayama A, et al. Differences in peritoneal solute transport rates in peritoneal dialysis. Clin Exp Nephrol 2019;23:122-34. [Crossref] [PubMed]
- Fischbach M, Schmitt CP, Shroff R, et al. Increasing sodium removal on peritoneal dialysis: applying dialysis mechanics to the peritoneal dialysis prescription. Kidney Int 2016;89:761-6. [Crossref] [PubMed]
- Vongsanim S, Davenport A. Factors associated with systolic hypertension in peritoneal dialysis patients. J Nephrol 2020;33:365-70. [Crossref] [PubMed]
- Ateş K, Nergizoğlu G, Keven K, et al. Effect of fluid and sodium removal on mortality in peritoneal dialysis patients. Kidney Int 2001;60:767-76. [Crossref] [PubMed]
- Sun T, Sakata F, Ishii T, et al. Excessive salt intake increases peritoneal solute transport rate via local tonicity-responsive enhancer binding protein in subtotal nephrectomized mice. Nephrol Dial Transplant 2019;34:2031-42. [Crossref] [PubMed]
- Shao H, Im H, Castro CM, et al. New Technologies for Analysis of Extracellular Vesicles. Chem Rev 2018;118:1917-50. [Crossref] [PubMed]
- Boriachek K, Islam MN, Möller A, et al. Biological Functions and Current Advances in Isolation and Detection Strategies for Exosome Nanovesicles. Small 2018;14: [Crossref] [PubMed]
- Thongboonkerd V. Roles for Exosome in Various Kidney Diseases and Disorders. Front Pharmacol 2019;10:1655. [Crossref] [PubMed]
- Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol 2018;141:1202-7. [Crossref] [PubMed]
- Grange C, Skovronova R, Marabese F, et al. Stem Cell-Derived Extracellular Vesicles and Kidney Regeneration. Cells 2019;8:1240. [Crossref] [PubMed]
- Carreras-Planella L, Soler-Majoral J, Rubio-Esteve C, et al. Proteomic profiling of peritoneal dialysis effluent-derived extracellular vesicles: a longitudinal study. J Nephrol 2019;32:1021-31. [Crossref] [PubMed]
- Corciulo S, Nicoletti MC, Mastrofrancesco L, et al. AQP1-Containing Exosomes in Peritoneal Dialysis Effluent As Biomarker of Dialysis Efficiency. Cells 2019;8:330. [Crossref] [PubMed]
- Kleyman TR, Kashlan OB, Hughey RP. Epithelial Na+ Channel Regulation by Extracellular and Intracellular Factors. Annu Rev Physiol 2018;80:263-81. [Crossref] [PubMed]
- Mutchler SM, Kleyman TR. New insights regarding epithelial Na+ channel regulation and its role in the kidney, immune system and vasculature. Curr Opin Nephrol Hypertens 2019;28:113-9. [Crossref] [PubMed]
- Iwai N, Baba S, Mannami T, et al. Association of a sodium channel alpha subunit promoter variant with blood pressure. J Am Soc Nephrol 2002;13:80-5. [Crossref] [PubMed]
- Valinsky WC, Touyz RM, Shrier A. Aldosterone, SGK1, and ion channels in the kidney. Clin Sci (Lond) 2018;132:173-83. [Crossref] [PubMed]
- Deng W, Li CY, Tong J, et al. Insulin ameliorates pulmonary edema through the upregulation of epithelial sodium channel via the PI3K/SGK1 pathway in mice with lipopolysaccharide-induced lung injury. Mol Med Rep 2019;19:1665-77. [Crossref] [PubMed]
- He J, Qi D, Tang XM, et al. Rosiglitazone promotes ENaC-mediated alveolar fluid clearance in acute lung injury through the PPARγ/SGK1 signaling pathway. Cell Mol Biol Lett 2019;24:35. [Crossref] [PubMed]
- Kim K, Hung RJ, Perrimon N. miR-263a Regulates ENaC to Maintain Osmotic and Intestinal Stem Cell Homeostasis in Drosophila. Dev Cell 2017;40:23-36. [Crossref] [PubMed]
- Morelle J, Stachowska-Pietka J, Öberg C, et al. ISPD recommendations for the evaluation of peritoneal membrane dysfunction in adults: Classification, measurement, interpretation and rationale for intervention. Perit Dial Int 2021;41:352-72. [Crossref] [PubMed]
- Trac PT, Thai TL, Linck V, et al. Alveolar nonselective channels are ASIC1a/α-ENaC channels and contribute to AFC. Am J Physiol Lung Cell Mol Physiol 2017;312:L797-811. [Crossref] [PubMed]
- Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018;7:1535750. [Crossref] [PubMed]
- Borrelli S, De Nicola L, Minutolo R, et al. Sodium toxicity in peritoneal dialysis: mechanisms and "solutions". J Nephrol 2020;33:59-68. [Crossref] [PubMed]
- Moreno JA, Hamza E, Guerrero-Hue M, et al. Non-Coding RNAs in Kidney Diseases: The Long and Short of Them. Int J Mol Sci 2021;22:6077. [Crossref] [PubMed]
- Wang Z, Zhou Z, Ji W, et al. Silencing of lncRNA 6030408B16RIK prevents ultrafiltration failure in peritoneal dialysis via microRNA-326-3p-mediated WISP2 down-regulation. Biochem J 2020;477:1907-21. [Crossref] [PubMed]
- Rigalli JP, Barros ER, Sommers V, et al. Novel Aspects of Extracellular Vesicles in the Regulation of Renal Physiological and Pathophysiological Processes. Front Cell Dev Biol 2020;8:244. [Crossref] [PubMed]
- Somiya M. Where does the cargo go?: Solutions to provide experimental support for the "extracellular vesicle cargo transfer hypothesis". J Cell Commun Signal 2020;14:135-46. [Crossref] [PubMed]
- Szeto CC, Chow KM, Kwan BC, et al. Peritoneal dialysis effluent miR-21 and miR-589 levels correlate with longitudinal change in peritoneal transport characteristics. Clin Chim Acta 2017;464:106-12. [Crossref] [PubMed]
- Chen J, Kam-Tao P, Kwan BC, et al. Relation between microRNA expression in peritoneal dialysis effluent and peritoneal transport characteristics. Dis Markers 2012;33:35-42. [Crossref] [PubMed]
- Xu T, Lei T, Li SQ, et al. DNAH17-AS1 promotes pancreatic carcinoma by increasing PPME1 expression via inhibition of miR-432-5p. World J Gastroenterol 2020;26:1745-57. [Crossref] [PubMed]
- Zhang J, Xu C, Gao Y, et al. A Novel Long Non-coding RNA, MSTRG.51053.2 Regulates Cisplatin Resistance by Sponging the miR-432-5p in Non-small Cell Lung Cancer Cells. Front Oncol 2020;10:215. [Crossref] [PubMed]
- Yoder JB, Ben-Johny M, Farinelli F, et al. Ca2+-dependent regulation of sodium channels NaV1.4 and NaV1.5 is controlled by the post-IQ motif. Nat Commun 2019;10:1514. [Crossref] [PubMed]
- Myers EJ, Yuan L, Felmlee MA, et al. A novel mutant Na+ /HCO3- cotransporter NBCe1 in a case of compound-heterozygous inheritance of proximal renal tubular acidosis. J Physiol 2016;594:6267-86. [Crossref] [PubMed]
- Rosenbaek LL, Rizzo F, MacAulay N, et al. Functional assessment of sodium chloride cotransporter NCC mutants in polarized mammalian epithelial cells. Am J Physiol Renal Physiol 2017;313:F495-504. [Crossref] [PubMed]
- Jalloul AH, Szerencsei RT, Rogasevskaia TP, et al. Structure-function relationships of K+-dependent Na+/Ca2+ exchangers (NCKX). Cell Calcium 2020;86:102153. [Crossref] [PubMed]
- De Paolis E, Scaglione GL, De Bonis M, et al. CYP24A1 and SLC34A1 genetic defects associated with idiopathic infantile hypercalcemia: from genotype to phenotype. Clin Chem Lab Med 2019;57:1650-67. [Crossref] [PubMed]
- Boscardin E, Alijevic O, Hummler E, et al. The function and regulation of acid-sensing ion channels (ASICs) and the epithelial Na(+) channel (ENaC): IUPHAR Review 19. Br J Pharmacol 2016;173:2671-701. [Crossref] [PubMed]
- Li XH, Hu YM, Yin GL, et al. Correlation between HCN4 gene polymorphisms and lone atrial fibrillation risk. Artif Cells Nanomed Biotechnol 2019;47:2989-93. [Crossref] [PubMed]
- Dobbs LG, Johnson MD. Alveolar epithelial transport in the adult lung. Respir Physiol Neurobiol 2007;159:283-300. [Crossref] [PubMed]
- Mutsaers SE. The mesothelial cell. Int J Biochem Cell Biol 2004;36:9-16. [Crossref] [PubMed]
- Nie HG, Tucker T, Su XF, et al. Expression and regulation of epithelial Na+ channels by nucleotides in pleural mesothelial cells. Am J Respir Cell Mol Biol 2009;40:543-54. [Crossref] [PubMed]


