Comprehensive genomic profiling and PD-L1 expression of primary lymphoepithelioma-like carcinoma of the stomach and parotid gland
Introduction
Lymphoepithelioma-like carcinoma (LELC) as an uncommon epithelial tumor has unique morphology and is characterized by irregular sheets, trabeculae, ill-defined tubules, or syncytia of polygonal cells embedded within a prominent lymphocytic infiltrate, with intraepithelial lymphocytes (1). It has been found in various anatomical sites, including the nasopharynx, larynx, tonsil, salivary gland, parotid gland, lung, breast, thymus, stomach, liver, duodenum, renal pelvis, urinary bladder, uterine cervix, endometrium, ovary, vulva, and vagina (2,3).
LELC of the stomach is a rare type of gastric cancer (GC), accounting for 1–4% of all GCs (4). It consists of two subsets, Epstein-Barr virus-positive (EBV+) and microsatellite instability-high (MSI-H) carcinomas (1,5). LELC of the stomach is closely associated with EBV, with 80–100% of cases being EBV-positive (6). The prevalence of MSI-H carcinoma in LELC of the stomach ranges from 7% to 39% (1,5). Previous studies have demonstrated that LELC of the stomach has a better prognosis than EBV+ and conventional GC (4,6,7). Radical resection is the standard of care for early-stage GC patients and advance-stage GC patients frequently receive palliative management, including programmed cell death-1 (PD-1)/programmed cell death ligand-1 (PD-L1) inhibitor and best supportive care (8). To date, there is no established agent for patients with advanced-stage LELC of the stomach due to the lack of genomic data. A number of studies on LELC of the stomach have been reported. However, it is commonly reported as case reports due to its rarity. In addition, the literature on its comprehensive genomic profiling is scant. To the knowledge, only one study has explored the molecular features of LELC of the stomach, which was performed in the Western population (9). However, its molecular features in the Chinese population have not been documented. In addition, the molecular features and clinicopathological characteristics of LELC of the parotid gland, which is a very rare malignancy, remain largely unknown. Surgical resection is the standard of care for patients with resectable salivary gland tumor. Definitive radiotherapy (RT) or concurrent systemic therapy and RT is the standard of care for patients with unresectable salivary gland tumor (10). Immune checkpoint inhibitors (ICIs) have promising efficacy and manageable safety in a variety of malignancies (11-13). Whether patients with LELC of the parotid gland have potential of utilizing ICI as a treatment regimen remains elusive. Moreover, the genomic profiling and the programmed cell death ligand-1 (PD-L1) expression status in LELC of the parotid gland has not been reported yet.
In the present study, we performed comprehensive genomic profiling and PD-L1 expression analysis in primary LELC of the stomach and parotid gland to investigate the molecular mechanisms of tumorigenesis and explore potential therapeutic strategies.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/atm-21-5908).
Methods
Patients
Thirty-five patients diagnosed with LELC who underwent surgical resection in Guangdong Provincial People’s Hospital between April 2015 and May 2020 were enrolled in this study. In the present work, LELC was defined as an epithelial tumor having irregular sheets, trabeculae, ill-defined tubules, or syncytia of polygonal cells embedded within a prominent lymphocytic infiltrate, with intraepithelial lymphocytes. All tumors were evaluated by two independent pathologists based on histopathological diagnosis from resected stomach lesion. Disease-free survival (DFS) was defined as the interval from the surgery date to the date of first tumor recurrence, distant metastasis, or death. Overall survival (OS) was defined as the interval from the surgery date to the date of death. Patients without an event were censored at the last follow-up visit. This study was approved by the Ethics Committee of Guangdong Provincial People’s Hospital (No. GDREC2018432A). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from each patient for the use of their tumor samples.
Tissue DNA isolation and library preparation
Tissue deoxyribonucleic acid (DNA) was extracted from formalin-fixed, paraffin-embedded tumor tissues (FFPE) with QIAamp DNA FFPE tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The concentration of DNA was measured using the Qubit 2.0 Fluorometer with Qubit double-stranded DNA assay kit (Life Technologies, Carlsbad, CA, USA). A minimum of 50 ng of DNA is required for next-generation sequencing (NGS) library construction. DNA extracted from tumors was fragmented using the Covaris M220 focused ultrasonicator (Covaris, Woburn, MA, USA), followed by end repair, phosphorylation, Diels-Alder addition, and adaptor ligation.
Capture-based targeted sequencing
Fragments with the size of 200–400 bp were selected using AMPure bead (Beckman Coulter Inc., Brea, CA, USA), followed by hybridization, hybrid selection, and polymerase chain reaction amplification. Samples were subsequently subjected to capture-based targeted sequencing using a panel consisting of 520 cancer-related genes (Burning Rock Biotech, Guangzhou, China). High sensitivity DNA electrophoresis was then performed using the Bioanalyzer system (Agilent Technologies Inc., Santa Clara, CA, USA) to assess the quality and size of the fragments. Indexed samples were sequenced on a Nextseq500 sequencer (Illumina, Inc., San Diego, CA, USA) with pair-end reads.
Sequence data analysis
The raw sequencing data were preprocessed using Trimmomatic 0.36 (14). Preprocessed sequencing data were then mapped to the human genome (hg19) using Burrows-Wheeler Aligner 0.7.10 (15). Variant calling and annotation were performed using GATK 3.2 (16), MuTect (17), and VarScan (18). According to the Exome Aggregation Consortium (ExAC, http://exac.broadinstitute.org), 1,000 Genomes, The Single Nucleotide Polymorphism Database (dbSNP, https://www.ncbi.nlm.nih.gov/snp/?term=), ESP6500SI-V2 database, variants with a population frequency >0.1% were defined as single nucleotide polymorphisms and excluded. The remaining variants were annotated with ANNOVAR (19) and SnpEff v3.6 (20). Copy number variations (CNVs) (including copy number amplification and deletion) and large genomic rearrangements (LGRs) were detected by in-house analysis scripts as previously described (21,22). DNA translocation analysis was performed using Tophat2 (23) and Factera 1.4.3 (24). The allele frequency (AF) of mutation was calculated. The MSI status of tumor sample was determined as previously described (25). Additionally, the tumor mutational burden (TMB) was analyzed, which was defined as the somatic single nucleotide variants (SNVs) and small insertions and deletions (InDels) located at the coding region and its 20 bp upstream/downstream region (26). TMB was calculated according to the following equation:
Immunohistochemistry staining analysis
Immunohistochemistry (IHC) staining analysis was conducted on FFPE tumor samples. All samples were pretreated and stained with the monoclonal mouse anti-PD-L1 antibody (clone number: 22C3, Dako, Agilent Technologies Inc., Santa Clara, CA, USA) and monoclonal mouse anti-cell differentiation (CD) 8 antibody (clone number: C8/144B, Gene tech, Shanghai, China) according to the manufacturer’s instruction. PD-L1 expression level was determined by combined positive score (CPS), which was computed as a ratio between the number of PD-L1-stained cells (tumor cells, lymphocytes, macrophages) and all viable cancer cells, multiplied by 100.
Statistical analysis
Differences in the groups were calculated and presented using Fisher’s exact test, paired two-tailed Student’s t-test, or analysis of variance (as appropriate). P<0.05 indicated statistical significance. All data were analyzed using R statistics package version 3.4.0 (Vienna, Austria). Correlations between somatic alterations and clinical outcomes were only investigated in patients with LELC of the stomach. Data used to support the results of this study can be obtained from the corresponding author.
Results
Patient characteristics
From April 2015 to May 2020, 35 patients consecutively diagnosed with LELC in Guangdong Provincial People’s Hospital were enrolled in our study, including 20 patients with LELC of the stomach, eight patients with LELC of the parotid gland, three patients with LELC of the liver, two patients with LELC of the salivary gland, one patient with LELC of the palate, and one patient with LELC of the orbit. Among the patients with LELC of the stomach, 17 (85.0%, 17/20) were males, 16 (80.0%, 16/20) had no smoking history, and the median age at diagnosis was 54.5 years, ranging from 25.0 to 81.0 years. The majority of patients with LELC of the stomach (80.0%, 16/20) were diagnosed at stage I–III. Among the patients with LELC of the parotid gland, seven (87.5%, 7/8) were males, all patients had no smoking history, and the median age at diagnosis was 31.0 years (ranging from 16.0 to 54.0 years). All cases were diagnosed at stage I–III. The clinical characteristics of patients are summarized in Table 1. All tumor samples collected from patients were EBV+.
Table 1
| Characteristics | LELC of the stomach | LELC of the parotid gland |
|---|---|---|
| Median age (range), years | 54.5 (25.0–81.0) | 31.0 (16.0–54.0) |
| Gender, n (%) | ||
| Female | 3 (15.0) | 1 (12.5) |
| Male | 17 (85.0) | 7 (87.5) |
| Smoking, n (%) | ||
| No | 16 (80.0) | 8 (100.0) |
| Yes | 2 (10.0) | 0 (0.0) |
| Unknown | 2 (10.0) | 0 (0.0) |
| Clinical stages, n (%) | ||
| Stage I | 3 (15.0) | 0 (0.0) |
| Stage II | 3 (15.0) | 4 (50.0) |
| Stage III | 10 (50.0) | 4 (50.0) |
| Stage IV | 3 (15.0) | 0 (0.0) |
| Unknown | 1 (5.0) | 0 (0.0) |
| Total | 20 | 8 |
LELC, lymphoepithelioma-like carcinoma.
Somatic alterations of LELC of the stomach
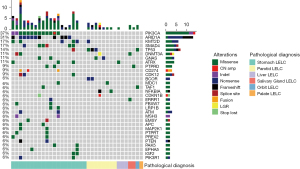
Capture-based targeted sequencing was performed on tumor tissue samples from 20 patients to elucidate the comprehensive mutational profiling of LELC of the stomach. Among the patients with LELC of the stomach, two had no somatic alterations detected from this panel (Figure 1). Collectively, 215 somatic alterations spanning 126 genes were identified from 18 patients with LELC of the stomach, including 183 SNVs, 23 InDels, two fusions, six CNVs, and one LGR. Each patient with LELC of the stomach harbored an average of 11.94 somatic alterations. The most frequently mutated genes included PIK3CA, ARID1A, SMAD4, and KMT2D, occurring in 65.0% (13/20), 55.0% (11/20), 30.0% (6/20), and 25.0% (5/20) of patients with LELC of the stomach, respectively (Figure 1). In addition, one patient harbored 9p24.1 region amplification (amplification of CD274, PDCD1LG2 [PD-L2], and JAK2) and two patients harbored CD274 fusion, including one patient with CD274-PLGRKT fusion and another patient with CD274-intergenic (CD274, PDCD1LG2) fusion.
Somatic alterations of LELC of parotid gland and other anatomical sites
Capture-based targeted sequencing was also performed on the tumor tissue samples from another 15 patients with LELC of different anatomical sites. Somatic alterations were detected in 7 patients with LELC of the parotid gland. A total of 37 somatic alterations spanning 30 genes, including 24 SNVs, five InDels, seven CNVs, and one LGR (Figure 1), were identified in seven patients with LELC of the parotid gland. Each patient with LELC of the parotid gland harbored an average of 5.29 somatic mutations. The most frequently mutated genes included TP53, GNAS, and BCOR, occurring in 37.5% (3/8), 25.0% (2/8), and 25.0% (2/8) of patients with LELC of the parotid gland, respectively (Figure 1).
In addition, somatic alterations were detected from this panel in three patients with LELC of the liver (average: 4.7 mutations), two patients with LELC of the salivary gland (average: 3.0 mutations), and one patient with LELC of the orbit (2.0 mutations). No somatic mutations were detected from this panel in the patient with LELC of the palate. Collectively, these findings indicated that patients with LELC of the stomach might carry a high burden of somatic alterations compared to those with LELC of other anatomical sites.
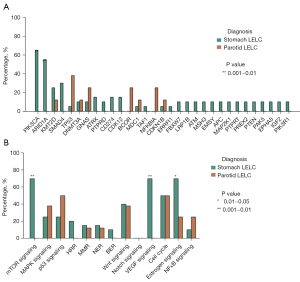
Comparison of genomic profiling between LELC of the stomach and parotid gland
Due to the limited cases of LELC of the liver, salivary gland, palate, and orbit, the genomic profiling of LELC of the stomach was only compared with that of LELC of the parotid gland in the present study. All LELC of the stomach/parotid gland samples were microsatellite-stable (MSS). Patients with LELC of the stomach had a higher mutation frequency in PIK3CA (65.0% vs. 0%, P=0.0025) and ARID1A (55.0% vs. 0%, P=0.0097), as well as a trend of lower mutation frequency in TP53 (5.0% vs. 38.0%, P=0.058) compared to those with LELC of the parotid gland (Figure 2A). Patients with LELC of the stomach also had a notably higher mutation frequency in the mechanistic target of rapamycin kinase (mTOR) (70.0% vs. 0%, P=0.0019) and vascular endothelial growth factor (VEGF) signaling pathway (70.0% vs. 0%, P=0.0019) compared to those with LELC of the parotid gland, respectively (Figure 2B).
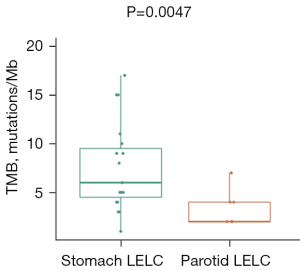
TMB of LELC of the stomach and parotid gland
The TMB level of patients with LELC of the stomach/parotid gland who underwent NGS was estimated. Eighteen and seven patients with LELC of the stomach and parotid gland had available TMB, respectively. The median TMB of patients with LELC of the stomach was 5.0 mutations/Mb (ranging from 1.0 to 21.9 mutations/Mb), which was 1.0 mutations/Mb (ranging from 1.0 to 6.0 mutations/Mb) in those with LELC of the parotid gland. LELC of the stomach showed a significantly increased TMB compared with LELC of the parotid gland (P=0.0047) (Figure 3).
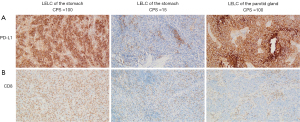
Immune microenvironment in patients with LELC of the stomach and parotid gland
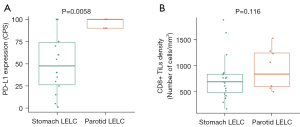
We also assessed the immune microenvironment in patients with LELC of the stomach/parotid gland by IHC staining for PD-L1 and CD8 who had adequate tumor samples, including 18 patients with LELC of the stomach and seven patients with LELC of the parotid gland. We restricted our analysis to CD8 positive (CD8+) tumor-infiltrating lymphocytes due to the fact that these cells are generally considered to be the main effector population following treatment with ICIs. Representative PD-L1 and CD8 staining is shown in Figure 4A,4B. PD-L1 expression level was assessed using the Dako 22C3 immunostaining assay. CPS was used to measure the PD-L1 expression level. Analysis revealed that all tumors had PD-L1 expression (CPS≥1). The median CPS of patients with LELC of the stomach was 47.5 (ranging from 1 to 100), which was 90 in patients with LELC of the parotid gland (ranging from 90 to 100). Compared with LELC of the parotid gland, LELC of the stomach showed a significantly decreased CPS (90.0 vs. 47.5, P=0.0058, Figure 5A). Moreover, CD8 immunostaining assay was performed in 19 and 8 patients with LELC of stomach and parotid gland, respectively. The density of CD8+ tumor infiltrating lymphocytes (TILs) in LELC of the stomach and parotid gland were comparable (717.9 vs. 707.1/mm2, P=0.116, Figure 5B).
Next, the correlation between PD-L1 expression and TMB was investigated in patients with LELC of the stomach/parotid gland. Our analysis showed no correlation between PD-L1 expression and TMB level with a Pearson correlation coefficient of −0.201 (P=0.334, Figure S1).
Correlations between somatic alterations and clinical outcomes in patients with LELC of the stomach
Due to the limited cases (n=5) of LELC of the parotid gland who had available survival data, the associations between genomic alterations and survival outcomes, including DFS and OS, were only evaluated in patients with LELC of the stomach. We found that the presence of recurrent PIK3CA, SMAD4, or KMT2D alterations was not associated with DFS or OS (Figure S2). Patients with SMAD4 alterations had a trend of shorter DFS than those without SMAD4 alterations. A comparable OS was also observed between patients with and without SMAD4 alterations.
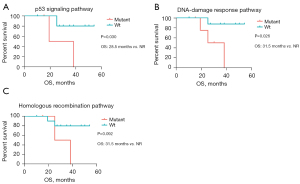
The associations between survival outcomes and 16 classic signaling pathways implicated in tumorigenesis were subsequently investigated (Table S1). Only those signaling pathways with somatic alterations detected in at least three patients were included for further analyses (Table S1). Six and seven signaling pathways were eventually included for DFS and OS analysis, respectively. We found that no signaling pathways were associated with DFS. However, patients with alterations in the p53 signaling pathway [28.5 months vs. not reached (NR), P=0.030, Figure 6A] and the DNA-damage response pathway (31.5 months vs. NR, P=0.026, Figure 6B) had a significantly shorter OS than those without alterations, respectively. Patients with alterations in the homologous recombination pathway showed a marginally shorter OS than those without alterations in this pathway (31.5 months vs. NR, P=0.092, Figure 6C).
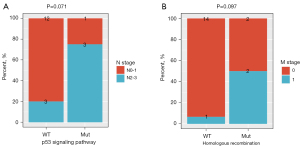
Next, we explored the associations between the presence of alterations in the p53 signaling pathway/homologous recombination pathway and the stage distribution in patients with LELC of the stomach. Our work revealed that patients with alterations in the p53 signaling pathway exhibited a trend of significant association with node metastasis (P=0.071, Figure 7A), and alterations in the homologous recombination pathway were marginally associated with the presence of distant metastasis (P=0.097, Figure 7B).
Discussion
To the best of our knowledge, this is the first study to comprehensively analyze the molecular features of LELC of the stomach in the Chinese population, and is the first to demonstrate the molecular features of LELC of the parotid gland. Our work might shed light on the molecular mechanism of tumorigenesis of these two rare malignancies.
LELC of the stomach is a rare histological type of GC, which is a heterogeneous disease with diverse molecular and histological subtypes. It has been documented that LELC of the stomach is male predominant (1). Similar results were observed in this work; the majority of patients (85%, 17/20) were males. We also found that LELC of the parotid gland was male predominant (87.5%, 7/8). Although a number of genomic analyses have been published on GC (27-32), very little consistent genomic alterations were observed across different individuals due to the high degree of heterogeneity among the GC of different subtypes. The Cancer Genome Atlas (TCGA) Research Network proposed a molecular classification that divides GC into four subtypes, one of which is EBV+ GC, displaying recurrent PIK3CA mutations, rare TP53 mutations, DNA hypermethylation, and amplification of JAK2, CD274, and PDCD1LG2 (32).
Consistent with previous studies (32,33), the presence of recurrent alterations in PIK3CA/CD274 and rare TP53 mutations in EBV+ GC was also observed in LELC of the stomach in this work (65%/15% and 5%). However, recurrent amplification at 9p24.1 (15%) containing JAK2, CD274, and PDCD1LG2 identified in EBV+ GC (32) was less commonly seen in EBV+ LELC of the stomach in this work (5%). In addition, tumor suppressors ARID1A, SMAD4, and KMT2D were also identified as recurrently mutated genes in LELC of the stomach.
The chromatin remodeling gene, ARID1A, has been reported to be a MSI- and EBV-associated putative driver gene (29,31), which is detected in 73% of EBV+ and 83% of MSI GCs (29). Genes in the transforming growth factor (TGF)-β pathway (SMAD2, SMAD4) are predicted to be key drivers in both MSI and MSS tumors (28). KMT2D, which encodes lysine methyltransferase 2D and has been reported as one of driver genes in GC, was identified as a frequently mutated gene in LELC of the stomach in the present work. We found that the frequency of ARID1A (55.0% vs. 53.8%, p=1.000), KTM2D (25.0% vs. 19.2%, P=0.726), and SMAD4 alterations (30.0% vs. 11.5%, P=0.149) between EBV+ LELC of the stomach and EBV+ GC from TCGA (32) was comparable, respectively. However, we did fail to detect ERBB2 amplification in any of our cases, although this feature is present in approximately 11.5% of EBV+ GC cases from TCGA and up to 34% of conventional GCs (34-36).
We also compared the mutation profiling of LELC of the stomach tumors from our cohort to the mutation landscape of EBV+ pulmonary LELC (PLELC) and EBV+ nasopharyngeal carcinoma (NPC) obtained from TCGA, which has been previously documented in the literature (3). PIK3CA and SMAD4 alterations were not observed in EBV+ PLELC/NPC. ARID1A (55.0% vs. 0% vs. 3.6%) and KMT2D-mutant frequency (25.0% vs. 0%. vs. 5.4%) of LELC of the stomach was significantly higher than that of EBV+ PLELC and EBV+ NPC. These findings suggest that the genomic profiling of EBV+ LELC of the stomach differs to EBV+ PLELC/NPC.
LELC of the parotid gland is much rarer than LELC of the stomach. To the best of our knowledge, it has been documented only in a few case reports (37,38). This study is the first to report the comprehensive genomic profiling of LELC of the parotid gland, and we found that TP53, GNAS, and BCOR were the most frequently mutated genes. We also compared the difference of recurrently mutated genes between LELC of the stomach and parotid gland; PIK3CA and ARID1A were identified as recurrently mutated genes in LELC of the stomach, but not in LELC of the parotid gland. Also, TP53 alterations were rare in LELC of the stomach, but were commonly seen in LELC of the parotid gland. These data revealed that the genomic profiling of LELC of the stomach was distinct from LELC of the parotid gland.
Due to its rarity, only one literature has reported on the molecular features of LELC of the stomach in a Western population (9), which demonstrates that LELC of the stomach is mutually exclusive EBV+ and mismatch repair deficiency (dMMR). However, it was closely associated with EBV in this work; all LELC of the stomach cases were EBV+ with MSS. A high ARID1A-mutant frequency and low TP53-mutant frequency were observed both in Chinese and Western LELC patients. In contrast, we detected PIK3CA alterations in 65% of LELC of the stomach cases in this study compared with only 25% of such cases in the Western population (9), and alterations in PRKDC, which encodes the catalytic subunit of DNA-dependent protein kinase involved in DNA double-strand break repair and recombination, were identified in 75% of cases in the Western population (9), but were not observed in this study. These data suggest that the genomic profiling of LELC of the stomach differs between Chinese and Western populations.
Several previous studies have documented that the prognosis of LELC of the stomach is better than that of EBV+ and conventional GC (4,6,7). Surgery is an important treatment option for early-stage LELC of the stomach. However, there is no established agent to treat patients with metastatic or recurrent LELC of the stomach. ICIs targeting the PD-/PD-L1 axis elicit promising efficacy in a variety of malignancies (39-41). Although the PD-L1 inhibitor pembrolizumab has been approved for the treatment of patients with recurrent locally advanced or metastatic GC whose tumors expressing PD-L1 with a CPS ≥1 [as determined by a Food and Drug Administration (FDA)-approved companion diagnostic test], the efficacy of pembrolizumab or other PD-1/PD-L1 inhibitor in LELC of the stomach remains elusive. EBV+ malignancies, including EBV+ GC and EBV+ NPC, have been demonstrated to be responsive to checkpoint immunotherapy (42,43). Several biomarkers have been reported to predict the efficacy of ICI, including the status of MSI/MMR, TMB, and PD-L1 expression (44,45). MSI status, TMB level, and PD-L1 expression level is generally believed to be independent of each other in solid tumors (46). In this context, we detected the MSI status, TMB level, and PD-L1 expression level in LELC of the stomach/parotid gland to preliminarily investigate whether PD-1/PD-L1 inhibitors could be treatment options for patients with LELC of the stomach or parotid gland. Vanderwalde and colleagues (47) have revealed that thirty percent of MSI-H cases were TMB-low, and only 26% of MSI-H are PD-L1 positive across cancer types. The similar results have been observed in the present work that all LELC of the stomach (n=18) or parotid gland (n=7) cases had an MSS tumor with PD-L1 expression, but only 6 LELC of the stomach cases had high TMB level (>7 mutations/Mb) and no LELC of the parotid gland cases had high TMB level. In the present work, we found no correlation between PD-L1 expression and TMB level in LELC patients (Pearson correlation coefficient =−0.201, P=0.334, Figure S1), which was consistent with previous studies indicating that PD-L1 expression level is not associated with TMB level in cancer patients (48-50). These findings suggest that MSI status, TMB level, and PD-L1 expression level were independent of each other in LELC of the stomach or parotid gland. Compared with LELC of the stomach, LELC of the parotid gland had a significantly higher expression level of PD-L1. Despite the low TMB and the presence of MSS in LELC of the stomach/parotid gland, the PD-L1 positivity of all tumors raises the potential of utilizing PD-1/PD-L1 inhibitor as a treatment regimen that could benefit these patients.
Generally believed, high PD-L1 expression is related to increased response rate and clinical benefit from PD-1/PD-L1 inhibitors (13,51). In the KYNOTE-062, pembrolizumab as monotherapy is not superior to chemotherapy in first-line, advanced gastric cancer patients with CPS of 1 or greater, but it prolongs OS vs. chemotherapy in patients with CPS of 10 or greater (52). Besides MSI status/TMB level/PD-L1 expression level, the presence of genomic alterations has been reported to be implicated in the response to PD-1/PD-L1 inhibitors. Alterations in POLE, EPHA, and NOTCH genes have been documented to be associated with favorable survival outcome in cancer patients who received PD-1/PD-L1 inhibitors. In the present work, 1, 2, and 0 LELC of the stomach patient(s) harbored alterations in POLE, EPHA, and NOTCH, respectively. No patients with LELC of the parotid gland were found to harbor alterations in POLE, EPHA, or NOTCH (53-55). The efficacy of PD-1/PD-L1 inhibitor has also been documented to be associated with the presence of driver mutations. In patients with oncogene-driven non-small cell lung cancer (NSCLC), PD-L1 expression level is significantly increased in the KRAS subgroup than in the non-KRAS subgroup and KRAS subgroup have a significantly higher objective response rate (ORR) than non-KRAS subgroup (16.1% vs. 5.7%) in those patients who received PD-1 inhibitor as monotherapy. Moreover, EGFR/ALK/ROS1-negative patients had a higher ORR than EGFR/ALK/ROS-positive patients in non-KRAS subgroup (10.5% vs. 0%) (56). Different ICIs display different efficacies in advanced cancer patients. For lung cancer, PD-1 inhibitor nivolumab (11,57) as monotherapy has been approved by both U.S. FDA and China National Medical Products Administration (NMPA) for the second-line rather than first-line treatment of the patients with advanced NSCLC. However, another PD-1 inhibitor pembrolizumab (58) as monotherapy has been approved by both FDA and NMPA for first-line treatment of patients with advanced NSCLC expressing PD-L1 (tumor proportion score ≥1%), with no EGFR or ALK genomic tumor aberrations. Clinical trials are needed to investigate the efficacy of each PD-1/PD-L1 inhibitor in LELC patients. In addition, clinical trials are warranted to determine whether the presence of alterations in POLE/EPHA/NOTCH or frequently mutated genes (such as PIK3CA, ARID1A) impacts on the efficacies of PD-1/PD-L1 inhibitors in LELC patients.
The p53 signaling pathway has been well-documented to contribute in cell cycle regulation, metabolism, aging and development, reproduction, suppression of tumor expression, and so on (59-61). Homologous recombination is essential for the accurate repair of DNA double-strand breaks (62,63). DNA-damage response is a complex signal transduction pathway that has the ability to sense DNA damage to influence cellular responses to DNA damage (64). Targeted therapy based on this principle, such as olaparib and the poly (ADP-ribose) polymerase (PARP) inhibitor, has been approved for treating patients with tumors lacking specific DNA-damage response functions, such as ovarian, breast, or prostate cancers with homologous recombination deficiencies (65). We found that alterations in the p53 signaling pathway and homologous recombination pathway were related to node and distant metastasis, respectively. In addition, alterations in these two signaling pathways and DNA-damage response predicted unfavorable prognosis in LELC of the stomach. These findings suggest that the p53 signaling pathway, homologous recombination pathway, and DNA-damage response pathway might play vital roles in tumorigenesis and serve as targets in the treatment of LELC of the stomach.
There are several limitations in this study that should be noted. Firstly, the associations between somatic alterations in signaling pathways and survival outcomes in patients with LELC of stomach were investigated. However, the censoring issue that the majority of patients were still alive at the end of follow-up might result in a bias in our conclusions. Secondly, there was a small sample size in this work, therefore, the molecular features of LELC of the stomach and parotid gland are needed to be investigated in a larger-scale, multicenter study.
Taken together, our findings reveal that the genomic landscape of LELC of the stomach is distinct to that of LELC of the parotid gland, and these two malignancies exhibit high PD-L1 expression. This study is the first to report the molecular features of LELC of the parotid gland, and is also the first to demonstrate the molecular features of LELC of the stomach in the Chinese patients, which were distinct from those in the Western population. The observation of high PD-L1 expression raises the potential of utilizing checkpoint immunotherapy for LELC of the stomach/parotid gland. A large, multicenter clinical trial study is needed to evaluate the treatment response of LELC of the stomach/parotid gland to PD-1/PD-L1 inhibitors.
Acknowledgments
Funding: This work was supported by the grants from the National Natural Science Foundation of China (No. 81802719), the Medical Scientific Research Foundation of Guangdong Province, China (No. A2017433), and the Research Grant of Guangdong Provincial People’s Hospital (Y02108141).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-5908
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-5908
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-5908). HD, TH, ML, YL, and ZZ are from Burning Rock Biotech, Guangzhou, China. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of Guangdong Provincial People’s Hospital (No. GDREC2018432A). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from each patient for the use of their tumor tissue samples.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cheng N, Hui DY, Liu Y, et al. Is gastric lymphoepithelioma-like carcinoma a special subtype of EBV-associated gastric carcinoma? New insight based on clinicopathological features and EBV genome polymorphisms. Gastric Cancer 2015;18:246-55. [Crossref] [PubMed]
- Hermel DJ, Du EZ, Lin R, et al. Checkpoint Inhibition in the Treatment of Unresectable, Advanced Lymphoepithelioma-like Hepatocellular Carcinoma. J Clin Transl Hepatol 2021;9:265-8. [Crossref] [PubMed]
- Xie Z, Liu L, Lin X, et al. A multicenter analysis of genomic profiles and PD-L1 expression of primary lymphoepithelioma-like carcinoma of the lung. Mod Pathol 2020;33:626-38. [Crossref] [PubMed]
- Watanabe H, Enjoji M, Imai T. Gastric carcinoma with lymphoid stroma. Its morphologic characteristics and prognostic correlations. Cancer 1976;38:232-43. [Crossref] [PubMed]
- Grogg KL, Lohse CM, Pankratz VS, et al. Lymphocyte-rich gastric cancer: associations with Epstein-Barr virus, microsatellite instability, histology, and survival. Mod Pathol 2003;16:641-51. [Crossref] [PubMed]
- Park S, Choi MG, Kim KM, et al. Lymphoepithelioma-like carcinoma: a distinct type of gastric cancer. J Surg Res 2015;194:458-63. [Crossref] [PubMed]
- Wang ZH, Zhao JJ, Yuan Z. Lymphoepithelioma-like gastric carcinoma: A case report and review of the literature. World J Gastroenterol 2016;22:3056-61. [Crossref] [PubMed]
- Network N C C. NCCN Clinical Practice Guidelines in Oncology (NCCN guidelines) Gastric Cancer Version 5.2021. 2021.
- Hissong E, Ramrattan G, Zhang P, et al. Gastric Carcinomas With Lymphoid Stroma: An Evaluation of the Histopathologic and Molecular Features. Am J Surg Pathol 2018;42:453-62. [Crossref] [PubMed]
- Network N C C. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) Head and Neck Cancers Version 3.2021. 2021.
- Carbone DP, Reck M, Paz-Ares L, et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2415-26. [Crossref] [PubMed]
- Shitara K, Özgüroğlu M, Bang YJ, et al. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): a randomised, open-label, controlled, phase 3 trial. Lancet 2018;392:123-33. [Crossref] [PubMed]
- Ding W, LaPlant BR, Call TG, et al. Pembrolizumab in patients with CLL and Richter transformation or with relapsed CLL. Blood 2017;129:3419-27. [Crossref] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [Crossref] [PubMed]
- Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11.10.1-11.10.33.
- do Valle ÍF, Giampieri E, Simonetti G, et al. Optimized pipeline of MuTect and GATK tools to improve the detection of somatic single nucleotide polymorphisms in whole-exome sequencing data. BMC Bioinformatics 2016;17:341. [Crossref] [PubMed]
- Koboldt DC, Chen K, Wylie T, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 2009;25:2283-5. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [Crossref] [PubMed]
- Cingolani P, Platts A. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80-92. [Crossref] [PubMed]
- Wu D, Xie Y, Jin C, et al. The landscape of kinase domain duplication in Chinese lung cancer patients. Ann Transl Med 2020;8:1642. [Crossref] [PubMed]
- Li YS, Jiang BY, Yang JJ, et al. Unique genetic profiles from cerebrospinal fluid cell-free DNA in leptomeningeal metastases of EGFR-mutant non-small-cell lung cancer: a new medium of liquid biopsy. Ann Oncol 2018;29:945-52. [Crossref] [PubMed]
- Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36. [Crossref] [PubMed]
- Newman AM, Bratman SV, Stehr H, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics 2014;30:3390-3. [Crossref] [PubMed]
- Zhu L, Huang Y, Fang X, et al. A Novel and Reliable Method to Detect Microsatellite Instability in Colorectal Cancer by Next-Generation Sequencing. J Mol Diagn 2018;20:225-31. [Crossref] [PubMed]
- Chen X, Fang L, Zhu Y, et al. Blood tumor mutation burden can predict the clinical response to immune checkpoint inhibitors in advanced non-small cell lung cancer patients. Cancer Immunol Immunother 2021;70:3513-24. [Crossref] [PubMed]
- Nagarajan N, Bertrand D, Hillmer AM, et al. Whole-genome reconstruction and mutational signatures in gastric cancer. Genome Biol 2012;13:R115. [Crossref] [PubMed]
- Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet 2014;46:573-82. [Crossref] [PubMed]
- Wang K, Kan J, Yuen ST, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet 2011;43:1219-23. [Crossref] [PubMed]
- Ge S, Li B, Li Y, et al. Genomic alterations in advanced gastric cancer endoscopic biopsy samples using targeted next-generation sequencing. Am J Cancer Res 2017;7:1540-53. [PubMed]
- Cui J, Yin Y, Ma Q, et al. Comprehensive characterization of the genomic alterations in human gastric cancer. Int J Cancer 2015;137:86-95. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Morales-Sanchez A, Fuentes-Panana EM. Epstein-Barr Virus-associated Gastric Cancer and Potential Mechanisms of Oncogenesis. Curr Cancer Drug Targets 2017;17:534-54. [Crossref] [PubMed]
- Gerson JN, Skariah S, Denlinger CS, et al. Perspectives of HER2-targeting in gastric and esophageal cancer. Expert Opin Investig Drugs 2017;26:531-40. [Crossref] [PubMed]
- Boku N. HER2-positive gastric cancer. Gastric Cancer 2014;17:1-12. [Crossref] [PubMed]
- Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet 2016;388:2654-64. [Crossref] [PubMed]
- Sinha P, Tanveer N, Bansal T, et al. Cytologic Clues to the Diagnosis of Lymphoepithelioma-Like Carcinoma of Parotid Gland with Possible Therapeutic Implications. Indian J Surg Oncol 2019;10:374-6. [Crossref] [PubMed]
- Mok MY, Shek WH, Wong RW. Lymphoepithelioma-like carcinoma of the parotid gland in a patient with rheumatoid arthritis. Clin Exp Rheumatol 2002;20:848-50. [PubMed]
- Peters S, Gettinger S, Johnson ML, et al. Phase II Trial of Atezolizumab As First-Line or Subsequent Therapy for Patients With Programmed Death-Ligand 1-Selected Advanced Non-Small-Cell Lung Cancer (BIRCH). J Clin Oncol 2017;35:2781-9. [Crossref] [PubMed]
- Kono K, Nakajima S, Mimura K. Current status of immune checkpoint inhibitors for gastric cancer. Gastric Cancer 2020;23:565-78. [Crossref] [PubMed]
- Kang YK, Boku N, Satoh T, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;390:2461-71. [Crossref] [PubMed]
- Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med 2018;24:1449-58. [Crossref] [PubMed]
- Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018;359:1350-5. [Crossref] [PubMed]
- Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol 2016;17:e542-51. [Crossref] [PubMed]
- Darvin P, Toor SM, Sasidharan Nair V, et al. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med 2018;50:1-11. [Crossref] [PubMed]
- Walk EE, Yohe SL, Beckman A, et al. The Cancer Immunotherapy Biomarker Testing Landscape. Arch Pathol Lab Med 2020;144:706-24. [Crossref] [PubMed]
- Vanderwalde A, Spetzler D, Xiao N, et al. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med 2018;7:746-56. [Crossref] [PubMed]
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633-41. [Crossref] [PubMed]
- Yu H, Chen Z, Ballman KV, et al. Correlation of PD-L1 Expression with Tumor Mutation Burden and Gene Signatures for Prognosis in Early-Stage Squamous Cell Lung Carcinoma. J Thorac Oncol 2019;14:25-36. [Crossref] [PubMed]
- Singal G, Miller PG, Agarwala V, et al. Association of Patient Characteristics and Tumor Genomics With Clinical Outcomes Among Patients With Non-Small Cell Lung Cancer Using a Clinicogenomic Database. JAMA 2019;321:1391-9. [Crossref] [PubMed]
- Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Shitara K, Van Cutsem E, Bang YJ, et al. Efficacy and Safety of Pembrolizumab or Pembrolizumab Plus Chemotherapy vs Chemotherapy Alone for Patients With First-line, Advanced Gastric Cancer: The KEYNOTE-062 Phase 3 Randomized Clinical Trial. JAMA Oncol 2020;6:1571-80. [Crossref] [PubMed]
- Bai H, Duan J, Li C, et al. EPHA mutation as a predictor of immunotherapeutic efficacy in lung adenocarcinoma. J Immunother Cancer 2020;8:e001315. [Crossref] [PubMed]
- Wang F, Zhao Q, Wang YN, et al. Evaluation of POLE and POLD1 Mutations as Biomarkers for Immunotherapy Outcomes Across Multiple Cancer Types. JAMA Oncol 2019;5:1504-6. [Crossref] [PubMed]
- Zhang K, Hong X, Song Z, et al. Identification of Deleterious NOTCH Mutation as Novel Predictor to Efficacious Immunotherapy in NSCLC. Clin Cancer Res 2020;26:3649-61. [Crossref] [PubMed]
- Ng TL, Liu Y, Dimou A, et al. Predictive value of oncogenic driver subtype, programmed death-1 ligand (PD-L1) score, and smoking status on the efficacy of PD-1/PD-L1 inhibitors in patients with oncogene-driven non-small cell lung cancer. Cancer 2019;125:1038-49. [Crossref] [PubMed]
- Bylicki O, Barazzutti H, Paleiron N, et al. First-Line Treatment of Non-Small-Cell Lung Cancer (NSCLC) with Immune Checkpoint Inhibitors. BioDrugs 2019;33:159-71. [Crossref] [PubMed]
- Mok TSK, Wu YL, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 2019;393:1819-30. [Crossref] [PubMed]
- Hu W, Feng Z, Teresky AK, et al. p53 regulates maternal reproduction through LIF. Nature 2007;450:721-4. [Crossref] [PubMed]
- Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol 2015;16:393-405. [Crossref] [PubMed]
- Tanikawa C, Zhang YZ, Yamamoto R, et al. The Transcriptional Landscape of p53 Signalling Pathway. EBioMedicine 2017;20:109-19. [Crossref] [PubMed]
- Wright WD, Shah SS, Heyer WD. Homologous recombination and the repair of DNA double-strand breaks. J Biol Chem 2018;293:10524-35. [Crossref] [PubMed]
- Sun Y, McCorvie TJ, Yates LA, et al. Structural basis of homologous recombination. Cell Mol Life Sci 2020;77:3-18. [Crossref] [PubMed]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell 2010;40:179-204. [Crossref] [PubMed]
- Peyraud F, Italiano A. Combined PARP Inhibition and Immune Checkpoint Therapy in Solid Tumors. Cancers (Basel) 2020;12:1502. [Crossref] [PubMed]
(English Language Editor: A. Kassem)