
Targeted next generation sequencing of circulating tumor DNA provides prognostic information for management in breast cancer patients
Introduction
Circulating tumor DNA (ctDNA), which are DNA fragments of tumor circulating in the blood, is known to be released from cancer cells into blood during various cell processes such as apoptosis and necrosis and from the tumor itself (1). Breast cancer is the most commonly diagnosed cancer in women (2), and the incidence of breast cancer patients in Korea has increased in recent years, with breast cancer showing the fifth highest cancer mortality rate (3). Previous studies showed that ctDNA is a potential biomarker for progression and may be indicative of a therapeutic response in breast cancer patients (4). ctDNA can be collected repeatedly and relatively un-invasively during regular follow up visits and thus can be an effective tool for monitoring the course of disease or predicting treatment efficacy.
High sensitivity is required for detection of ctDNA due to the low presence of ctDNA in plasma and a high signal-to-noise ratio (5,6). Polymerase chain reaction (PCR)-based assays or next-generation sequencing (NGS)-based assays are performed to detect ctDNA (7), and NGS is a powerful tool in molecular screening programs because it can detect somatic mutations at quantities below 5% and ctDNA mutations in small amounts (8,9). It is known that there is a background error of 0.1% due to PCR, cluster generation, and sequencing processes in standard NGS analysis. The molecular barcoding system can reduce errors through the following technical processes. Tagging a unique molecular index (UMI) to cfDNA extracted from plasma. Then, the library preparation and sequencing process are carried out. The produced sequence is sorted by UMI and grouped into a family. A consensus sequence is created from the family sequences. If the same type of variant exists at the same location in all sequences, it can be considered as a real variant, and independent type variants that exist in each sequence can be treated as noises. Creating a consensus sequence is a key process, and it is known that errors can be reduced by about 100–100,000,000 times (10,11). By lowering the error, more sensitive analysis is possible. This is advantageous when analyzing samples with low tumor burden such as liquid biopsy.
Although there have been numerous studies on disease diagnosis and monitoring progression using NGS of ctDNA in breast cancer patients, there is a need for further study of ctDNA analysis in the practical clinical setting. The aim of this study was to assess the correlation between ctDNA and tumor DNA in breast cancer patients and evaluate the clinical utility of ctDNA as a therapeutic marker. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-4881/rc).
Methods
Patients and sample collection
The study recruited a total of 38 breast cancer patients, who all provided informed consent, at the National Cancer Center in Korea from August 2016 to July 2018 and was approved by Institutional Review Board of the National Cancer Center in Korea (IRB No. NCC2016-0202, NCC2016-0221, NCC2016-0272). All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Blood samples were collected before surgery or after chemotherapy and during follow-up. In total, 20 preserved formalin-fixed paraffin-embedded (FFPE) and fresh frozen (FF) tissue samples were obtained from the biobank of the National Cancer Center, Korea. Fresh frozen tissue samples are confirmed by anatomic pathologists for tumor proportion prior to banking. Of the 20 tissue samples, 14 were of breast tissue (primary or relapsed) and 6 were from metastatic sites.
Immunohistochemistry of tissue sections
Immunohistochemical (IHC) staining was performed on tissue sections cut from formalin-fixed, paraffin-embedded representative breast tumors. Staining was performed with Ventana ES autostainer (Ventana Medical Systems, Tucson, AZ, USA), using primary antibodies against ER (Ventana Medical Systems), PR (Ventana Medical Systems) and C-erbB2 (Ventana Medical Systems).
Extraction of ctDNA from plasma and genomic DNA from tissue
Blood samples were processed within 2 hours after collection. The samples were centrifuged at 3,000 rpm for 10 min at 4 °C and then the supernatant was centrifuged again (10 min at 16,000 ×g and 4 °C) to remove any remaining contaminating cells. ctDNA was extracted using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) from 2 mL of plasma according to the manufacturer’s instructions. ctDNA samples were quantified using the Qubit dsDNA HS (High Sensitivity) Assay Kit (Life Technologies, Carlsbad, CA, USA). Genomic DNA was extracted from 1 mL of whole blood with the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Tumor DNA was isolated from FFPE tissue and fresh tissue using the Qiagen AllPrep DNA/RNA FFPE kit.
Library preparation and sequencing
The ctDNA samples were amplified using the Oncomine Breast cfDNA Assay v2, which covers single nucleotide variations and mutations in AKT1, EGFR, ERBB3, ESR1, KRAS, PIK3CA, TP53, FBXW7, SF3B1 and copy number alterations in CCND1, ERBB2, FGFR1. The resulting libraries were quantified using the Ion Library TaqMan® Quantitation Kit (Thermo Fisher, Waltham, MA, USA). The prepared libraries were then sequenced on an Ion S5 XL Sequencer using the Ion 530™ kit and Ion 540™ kit (Thermo Fisher, Waltham, MA, USA). Somatic variants were identified using Sanger sequencing for allele mutation frequencies ≥30%. The Catalogs of Somatic Mutations in Cancer (COSMIC), ClinVar, and dbSNP were used to identify somatic variants. Data analysis was done via Oncomine TagSeq Breast v2 Liquid Biopsy 2.0 default options with minimum molecular cutoff of 2 and minimum mutant allele frequency of 0.05% (minimum variant molecular count – 0.5/molecular coverage). Whole exome sequencing (WES) was performed on matched tumor DNA (n=20).
Analysis of follow-up samples with droplet digital PCR
Mutations in extracted ctDNA were detected by droplet digital PCR (ddPCR) on a QX200 Droplet Digital PCR System (Bio-Rad Laboratories, Hercules, CA, USA). Each probe assay was obtained from Bio-Rad: AKT1 p.E17K, ERBB2 p.V842I, KRAS p.G12D, p.G12V, PIK3CA p.H1047R, p.E542K, TP53 p.R175H, p.R196*, p.Y220C, p.R306* and WT accordingly. ESR1 p.D538G, p.E380Q, p.Y537N, and p.Y537S probe assays were ordered from Life Technologies (Thermo Fisher, Waltham, MA, USA). Analyses were performed by Quanta-Soft software (Bio-Rad Laboratories, Hercules, CA, USA). The limit of detection (LOD) was confirmed by ddPCR with serially diluted DNA to 50%, 10%, 1%, 0.5%, 0.25%, 0.1%, 0.05% and 0.01% using wild type and mutant DNA.
Measurement of serum tumor markers
Serum concentrations of carcinoembryonic antigen (CEA) and CA15-3 were measured by chemiluminescent microparticle immunoassay with an Architect i2000SR Immunoassay Analyzer (Abbott Laboratories, Chicago, IL, USA) with the median cut-off value of <5.0 ng/mL and <31.3 U/mL, respectively. Serum HER2 was measured by ADVIA Centaur XP (Siemens Diagnostics, Tarrytown, NY, USA) with the median cutoff value of <15.0 ng/mL.
Statistical analyses
Statistical analyses were performed with GraphPad Prism 5.0 software (GraphPad Software Inc., La Jolla, CA, USA) and MedCalc for Windows, version 19.6 (MedCalc Software, Ostend, Belgium). The patient survival curves were calculated using the Kaplan-Meier (KM) method and the log-rank test. Multivariate analysis was done using Cox Regression for evaluating the effect of subtype and mutation status. Progression-free survival (PFS) was measured from the day of diagnosis to the day of progression or death, and overall survival (OS) was calculated from the day of diagnosis to the day of last follow-up or death. The effects of variants detected in the panel on OS or PFS were presented as hazard ratios (HR) with a 95% confidence interval (CI).
Results
Characteristics of breast cancer patients
The characteristics of the 38 patients included in the study are summarized in Table 1. The median age of the study subjects was 47 (range, 30–65). The subtypes based on immunohistochemistry (IHC) found at the time of diagnosis were triple-negative breast cancer (TNBC), hormone receptor (HR)+/human epidermal growth factor receptor 2 (HER2)−, HR−/HER2+, HR+/HER2+, and the number of patients corresponding to each subtype were 10 (26%), 23 (60%), 4 (11%) and 1 (3%), respectively. All patients except for one were at stage IV at the time blood was drawn for ctDNA analysis and the metastatic organs are listed in Table 1. Some patients had multiple metastatic organs. There were no statistically significant differences in age and immunohistochemical subtypes between the patient group with variants detected in ctDNA and the group without. ctDNA was also detected in luminal type showing HR+ and HER2− which is known to be of low-grade with good prognosis.
Table 1
| Characteristics | Detected | Not detected | P value |
|---|---|---|---|
| ctDNA | 0.555 | ||
| No. of patients [%] | 31 [82] | 7 [18] | |
| Median age [range], years | 46 [30–65] | 44 [32–62] | |
| Subtype [%] | 0.946 | ||
| TNBC | 8 [26] | 2 [29] | |
| HR+/HER2− | 19 [61] | 4 [57] | |
| HR−/HER2+ | 3 [10] | 1 [14] | |
| HR+/HER2+ | 1 [3] | 0 [0] | |
| Treatments other than chemotherapy [%] | 0.932 | ||
| Aromatase inhibitor | 18 [58] | 4 [57] | |
| Tamoxifen | 10 [32] | 2 [29] | |
| Herceptin | 3 [10] | 1 [14] | |
| Metastatic organs | 0.074 | ||
| Bone, bone marrow, spine | 11 | 1 | |
| Liver | 8 | 2 | |
| Brain | 7 | 1 | |
| Lung | 4 | 2 | |
| Lymph node | 0 | 2 | |
| Soft tissue | 1 | 0 |
TNBC, triple-negative breast cancer; HR, hormone receptor; HER2, human epidermal growth factor receptor 2.
Detection of somatic single nucleotide variants and copy number alterations in ctDNA
Somatic single nucleotide variants and copy number alterations (CNA) were detected in 31 (82%) of 38 patients, including 86 variants and 9 CNAs. Variants were most commonly identified in TP53 (50%), PIK3CA (15%) and ESR1 (14%) (Figure 1). The types of mutation for each gene are also shown (Table 2). ESR1 mutations were exclusively detected in ctDNA only and aromatase inhibitor was used in 7 of 8 (87.5%) positive patients. Aromatase inhibitor was used in 22 patients and among them 7 (31.8%) patients showed ESR1 mutations. In 4 patients (4/8, 50%) more than two types of mutations in ESR1 was found, with frequency of mutation in the following order, p.D538G (6/13, 46%), p.Y537S (3/13, 23%), p.Y537N (2/13, 15%), p.Y537C (1/13, 7%) and pE380Q (1/13, 7%) (Table 2).
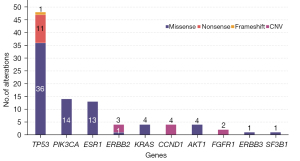
Table 2
| Gene | AA mutation | Mutations type | Frequency |
|---|---|---|---|
| TP53 | p.R306* | Nonsense | 9/48 |
| p.E286G | Missense | 5/48 | |
| p.Y220C | Missense | 5/48 | |
| p.R280K | Missense | 2/48 | |
| p.R248Q | Missense | 2/48 | |
| p.R273C | Missense | 2/48 | |
| p.R273H | Missense | 2/48 | |
| p.Q192* | Nonsense | 2/48 | |
| p.V272M | Missense | 2/48 | |
| p.M133K | Missense | 1/48 | |
| p.R175H | Missense | 1/48 | |
| p.C176F | Missense | 1/48 | |
| p.H179R | Missense | 1/48 | |
| p.A189V | Missense | 1/48 | |
| p.H193R | Missense | 1/48 | |
| p.H214R | Missense | 1/48 | |
| p.V216M | Missense | 1/48 | |
| p.P219S | Missense | 1/48 | |
| p.P222S | Missense | 1/48 | |
| p.R248W | Missense | 1/48 | |
| p.R273L | Missense | 1/48 | |
| p.P278L | Missense | 1/48 | |
| p.P278S | Missense | 1/48 | |
| p.R282W | Missense | 1/48 | |
| p.E286K | Missense | 1/48 | |
| p.Q331fs | Frameshift | 1/48 | |
| PIK3CA | p.H1047R | Missense | 5/14 |
| p.H1047L | Missense | 2/14 | |
| p.E545K | Missense | 2/14 | |
| p.E542K | Missense | 1/14 | |
| p.E726K | Missense | 1/14 | |
| p.M1043I | Missense | 1/14 | |
| p.N345K | Missense | 1/14 | |
| p.Q546K | Missense | 1/14 | |
| ESR1 | p.D538G | Missense | 6/13 |
| p.Y537S | Missense | 3/13 | |
| p.Y537N | Missense | 2/13 | |
| p.Y537C | Missense | 1/13 | |
| p.E380Q | Missense | 1/13 | |
| KRAS | p.G12V | Missense | 2/4 |
| p.G12S | Missense | 1/4 | |
| p.G12D | Missense | 1/4 | |
| AKT1 | p.E17K | Missense | 4/4 |
| ERBB2 | p.L755S | Missense | 1/1 |
| ERBB3 | p.E928G | Missense | 1/1 |
| SF3B1 | p.K700E | Missense | 1/1 |
*, nonsense mutation.
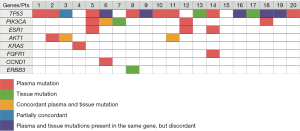
Concordance of detected variants between ctDNA and tumor DNA
Detected variants were compared in 20 patients who had results of tumor DNA and ctDNA. The most frequently detected gene alterations in tumor DNA were in TP53 (39%), PIK3CA (15%), and MUC16 (10%). The concordance on positives was defined as the detection of single nucleotide variants in both ctDNA and tumor DNA at the same gene location (Figure 2), and of 31 detected variants 3 (9.7%) showed concordant variance. The median time interval between tissue and blood collection was 1 month (range, 0–40 months).
Implications on prognosis in relation to the detected variants in ctDNA
Survival was analyzed and compared between patients with and without somatic mutations in each gene. There was no statistically significant difference in PFS and OS between patients with and without somatic mutations in PIK3CA and ESR1. However, patients with mutations in TP53 showed significantly worse OS compared to those without [hazard ratio (HR) =3.90, 95% confidence interval (CI): 1.10–13.84, P=0.035] (Figure 3). Cox regression analysis of hormonal subtype and gene mutation detected in ctDNA was done for PFS and OS and triple negative breast cancer (TNBC) (HR =8.44, 95% CI: 1.50–47.47, P=0.016) and TP53 mutation (HR =6.45, 95% CI: 1.13–36.83, P=0.036) showed to be statistically significant worse prognosis factor for OS (Table 3). Patients with TP53 mutations showed high prevalence of leptomeningeal involvement, 6 of 7 patients (86%) which may have contributed to high hazard ratio for OS.
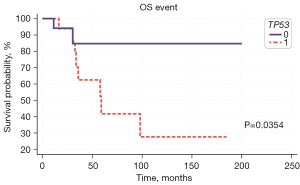
Table 3
| Variable | PFS | OS | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Subtype | |||||||
| TNBC | 1.69 | 0.70 to 4.09 | 0.247 | 8.44 | 1.50 to 47.47 | 0.016 | |
| Detected mutation (ctDNA) | |||||||
| TP53 | 0.941 | 0.44 to 1.99 | 0.875 | 6.45 | 1.13 to 36.83 | 0.036 | |
| PIK3CA | 1.08 | 0.47 to 2.48 | 0.862 | 1.02 | 0.23 to 4.63 | 0.976 | |
| ESR1 | 0.53 | 0.19 to 1.43 | 0.210 | 0.91 | 0.16 to 5.10 | 0.919 | |
TNBC, triple negative breast cancer; PFS, progression free survival; OS, overall survival; ctDNA, circulating tumor DNA; HR, hazard ratio; CI, confidence interval.
Serial monitoring of somatic single nucleotide variants in ctDNA
We performed serial monitoring of somatic single nucleotide variants in ctDNA with ddPCR in 5 patients. In cases where extracted DNA from tumor tissue was available, ddPCR was performed and the same mutations were detected in different quantities (data not shown).
Patient 5 was diagnosed with TNBC in March 2017. She received neoadjuvant therapy and in August same year, curative modified radical mastectomy (MRM) was performed and tumor tissue and blood were collected. She was the only one person whose stage was 2 when blood for ctDNA was drawn. The TP53 p.Y220C variant was detected in blood ctDNA using Oncomine panel [variant allele frequency (VAF) 9.25%] and ddPCR (VAF 4.40%). The patient was treated with capecitabine as adjuvant therapy and underwent radiation therapy (RT). In the second collection, after 10 months, ctDNA VAF decreased to 0.50% (Figure 4A) and there was no evidence of tumor in computed tomography (CT) image.
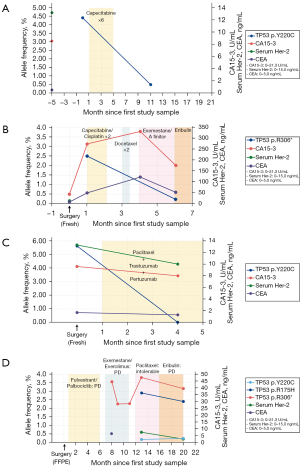
Patient 8 (ER positive, PR positive, HER2 negative) was diagnosed with metastatic breast cancer involving skin and axillary lymph node. The variants were not detected in tumor tissues in February 2018. One month later, TP53 p.R306* (VAF 2.5%) was detected in ctDNA and tumor markers such as CA15-3 and CEA were elevated. During the follow up, treatment regimens were switched for there was no response. After 6 months, the VAF of mutations in ctDNA decreased to 0.25% together with CA15-3 and CEA and CT findings revealed slight decrease in the tumor and infiltration (Figure 4B).
Patient 9 (ER negative, PR negative, HER2 positive) was diagnosed with breast cancer with bone metastasis in May 2014. After receiving neoadjuvant chemotherapy, palliative MRM was performed in July, 2018 and tumor tissue and blood was collected. TP53 p.Y220C variant was detected in blood ctDNA using Oncomine panel (VAF 5.90%) and ddPCR (VAF 5.63%). Patient received therapy of paclitaxel, trastuzumab, pertuzumab and is in stable disease status with no change in multiple bone metastasis. The values of tumor marker decreased at 4 months and ctDNA was not detected (Figure 4C). Image findings of metastasized bone lesions did not disappear in short time, however, ctDNA showed swift disappearance.
Patient 10 (ER positive, PR positive, HER2 negative) was diagnosed with stage I breast cancer in October 2013 and received curative mastectomy, adjuvant chemotherapy and endocrine therapy. TP53 p.R175H mutation (VAF 37.96%) was detected in tumor tissue. The tumor recurred in December 2015 in lymph node and she received neoadjuvant therapy and another surgery. However, the tumor relapsed in liver in November 2016. She received therapy of fulvestrant combined with palbociclib but she had intolerable adverse reactions and had to change treatment regimens. The image findings suggested increase in size and number of multiple liver metastases when the variants of TP53 p.Y220C (VAF 0.08%), p.R175H (VAF 2.43%) and p.R306* (VAF 3.16%) were detected in the ctDNA (Figure 4D).
Discussion
This study shows the results of ctDNA detected using a commercial NGS panel in breast cancer patients. In this study, variants of TP53, PIK3CA and ESR1 were the most frequently detected. Previous reports have shown that TP53, PIK3CA, ERBB2 and KRAS variants were most commonly identified in ctDNA panels from breast cancer patients (12-14). In our study, the detection rate of variants at the same position in both ctDNA and tumor DNA was 9.7%. Similar to our study, a study by Chae et al. reported a concordance rate on positives of 10.8% (13). The reason for low concordance rate of positive variants between tumor and ctDNA was the long interval between tumor and blood sample collection (median: 1 month; range, 0–40 months). Positive concordance in patients with intervals between tissue and blood sampling of less than 10 months was higher than in those with intervals of 10–30 months (data not shown). In another study, among 50 lung cancer patients, those with an interval ≤2 weeks (100%) showed higher concordance than those with an interval >6 months (60%) (15). There are many explanations for the discordance, such as intratumor heterogeneity, subclones within a primary tumor (16-18) and ctDNA arising from multiple metastatic sites. Additionally, ctDNA assays only identify mutations after tumor cells outgrow the blood supply, become hypoxic and undergo apoptosis or necrosis (19). It has been reported that mutations in the ESR1 gene, which encodes for the estrogen receptor (ER), arise as a result of chronic exposure to hormonal blockade during the adjuvant or the metastatic setting (20-22), and these mutations are virtually undetectable in primary tumors (23-25). These mutations lead to hyperactivation of the ER signaling system and are linked to adverse disease course, and ESR1 mutations detected in cell-free DNA (cfDNA) were associated with more aggressive disease biology in the BOLERO-2 clinical trial (26). In our study all ESR1 mutations were detected only by ctDNA analysis and 7 of 8 patients with ESR1 mutations had history of using aromatase inhibitor.
The sequencing depth, gene frequency threshold and gene coverage position may also be the reason for the low concordance between matched tumor DNA and ctDNA. In our study, the mutations detected in ctDNA was confirmed by ddPCR done on extracted DNA from tumor tissue but not by WES, reflecting the difference of sensitivity as the reason for discrepancy. The LOD of WES using tumor DNA was 5% and the depth was 200×. The LOD of the Oncomine Breast cfDNA panel was 0.05–0.35%, and the mean read depth was 39,704×. Since ctDNA panels have lower LOD than WES using tumor DNA, more variants could be detected. Several previous studies have compared hotspot gene positions and showed high concordance between ctDNA and tumor DNA. A comparison of exon 19 deletion and L858R EGFR mutation in non-small cell lung cancer (NSCLC) patients showed a high agreement of 80–98% (27). In a study of prostate cancer, there was 89% agreement between ctDNA and tumor DNA for 9 genes, including the driver genes AR, BRCA2, and ATM (28). In breast cancer, tissue DNA and ctDNA showed high agreement for PIK3CA mutation and ERBB2 amplifications (29). With respect to our study, the panel includes the position of a broader range of genes as well as hotspots and this could be the explanation for the lower concordance rate.
Patients with mutations in TP53 showed poorer OS than patients without. In a previous study, patients without mutations in the solid tumor had a better OS than those with (HR =0.26, 95% CI: 0.1409–0.9520, P<0.04) (6). In other studies, TNBC and early breast cancer patients with variants in ctDNA had significantly shorter disease-free survival (DFS) than patients without (30,31). In our study, TNBC subtype and TP53 mutation in ctDNA were related to statistically significant poor OS in multivariate analysis. To analyze the usefulness of the ctDNA, we monitored the changes of VAF according to therapeutic response. In our study, the PIK3CA and TP53 genes were monitored. The variants concurrently increased in patients with progressive disease, and the variants decreased in patients who underwent radiation therapy. The change in mutant allele frequency was also consistent with the tumor marker measurements; however, several mutations were detected at a value below the ddPCR LOD and monitoring was done in a small number of patients. There are many limitations of this study. Although most patients were at stage IV at the time blood was collected the patients had received different treatments and was not in a strictly designed clinical setting. And we can not rule out clonal hematopoiesis of indeterminate potential (CHIP) because we did not evaluate the gDNA mutation, even though evaluated genes are breast cancer specific genes and rarely reported as CHIP genes except TP53.
In summary, we have shown that the genomic profile of ctDNA in breast cancer patients provides additional prognostic information to the tumor DNA genome profile. TP53 mutations in detected in ctDNA was associated with poorer OS. In future studies, we need to monitor a larger number of patients to assess the clinical utility in actual practice. In addition, we need to characterize specific abnormal variants in ctDNA that can be used as prognostic markers and markers of therapeutic response in breast cancer patients.
Acknowledgments
Funding: This study was funded by the National Cancer Center, Korea (Grant No. 1611161).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-4881/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-4881/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-4881/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The study was approved by Institutional Review Board of the National Cancer Center in Korea (IRB No. NCC2016-0202, NCC2016-0221, NCC2016-0272). Informed consent was obtained from all individual participants included in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Jung KW, Won YJ, Kong HJ, et al. Prediction of Cancer Incidence and Mortality in Korea, 2018. Cancer Res Treat 2018;50:317-23. [Crossref] [PubMed]
- Madhavan D, Wallwiener M, Bents K, et al. Plasma DNA integrity as a biomarker for primary and metastatic breast cancer and potential marker for early diagnosis. Breast Cancer Res Treat 2014;146:163-74. [Crossref] [PubMed]
- Lupini L, Moretti A, Bassi C, et al. High-sensitivity assay for monitoring ESR1 mutations in circulating cell-free DNA of breast cancer patients receiving endocrine therapy. Sci Rep 2018;8:4371. [Crossref] [PubMed]
- Mehrotra M, Singh RR, Loghavi S, et al. Detection of somatic mutations in cell-free DNA in plasma and correlation with overall survival in patients with solid tumors. Oncotarget 2017;9:10259-71. [Crossref] [PubMed]
- Zill OA, Greene C, Sebisanovic D, et al. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. Cancer Discov 2015;5:1040-8. [Crossref] [PubMed]
- Page K, Guttery DS, Fernandez-Garcia D, et al. Next Generation Sequencing of Circulating Cell-Free DNA for Evaluating Mutations and Gene Amplification in Metastatic Breast Cancer. Clin Chem 2017;63:532-41. [Crossref] [PubMed]
- Risberg B, Tsui DWY, Biggs H, et al. Effects of Collection and Processing Procedures on Plasma Circulating Cell-Free DNA from Cancer Patients. J Mol Diagn 2018;20:883-92. [Crossref] [PubMed]
- Schmitt MW, Kennedy SR, Salk JJ, et al. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A 2012;109:14508-13. [Crossref] [PubMed]
- Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9:eaan2415. [Crossref] [PubMed]
- Frenel JS, Carreira S, Goodall J, et al. Serial Next-Generation Sequencing of Circulating Cell-Free DNA Evaluating Tumor Clone Response To Molecularly Targeted Drug Administration. Clin Cancer Res 2015;21:4586-96. [Crossref] [PubMed]
- Chae YK, Davis AA, Jain S, et al. Concordance of Genomic Alterations by Next-Generation Sequencing in Tumor Tissue versus Circulating Tumor DNA in Breast Cancer. Mol Cancer Ther 2017;16:1412-20. [Crossref] [PubMed]
- Maxwell KN, Soucier-Ernst D, Tahirovic E, et al. Comparative clinical utility of tumor genomic testing and cell-free DNA in metastatic breast cancer. Breast Cancer Res Treat 2017;164:627-38. [Crossref] [PubMed]
- Thompson JC, Yee SS, Troxel AB, et al. Detection of Therapeutically Targetable Driver and Resistance Mutations in Lung Cancer Patients by Next-Generation Sequencing of Cell-Free Circulating Tumor DNA. Clin Cancer Res 2016;22:5772-82. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Hiley C, de Bruin EC, McGranahan N, et al. Deciphering intratumor heterogeneity and temporal acquisition of driver events to refine precision medicine. Genome Biol 2014;15:453. [Crossref] [PubMed]
- Yates LR, Gerstung M, Knappskog S, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 2015;21:751-9. [Crossref] [PubMed]
- Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659-65. [PubMed]
- Jeselsohn R, Yelensky R, Buchwalter G, et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res 2014;20:1757-67. [Crossref] [PubMed]
- Robinson DR, Wu YM, Vats P, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet 2013;45:1446-51. [Crossref] [PubMed]
- Toy W, Shen Y, Won H, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet 2013;45:1439-45. [Crossref] [PubMed]
- Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012;486:400-4. [Crossref] [PubMed]
- Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333-9. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61-70. [Crossref] [PubMed]
- Chandarlapaty S, Chen D, He W, et al. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol 2016;2:1310-5. [Crossref] [PubMed]
- Zhu G, Ye X, Dong Z, et al. Highly Sensitive Droplet Digital PCR Method for Detection of EGFR-Activating Mutations in Plasma Cell-Free DNA from Patients with Advanced Non-Small Cell Lung Cancer. J Mol Diagn 2015;17:265-72. [Crossref] [PubMed]
- Wyatt AW, Annala M, Aggarwal R, et al. Concordance of Circulating Tumor DNA and Matched Metastatic Tissue Biopsy in Prostate Cancer. J Natl Cancer Inst 2017;109:djx118. [Crossref] [PubMed]
- Liang DH, Ensor JE, Liu ZB, et al. Cell-free DNA as a molecular tool for monitoring disease progression and response to therapy in breast cancer patients. Breast Cancer Res Treat 2016;155:139-49. [Crossref] [PubMed]
- Chen YH, Hancock BA, Solzak JP, et al. Next-generation sequencing of circulating tumor DNA to predict recurrence in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy. NPJ Breast Cancer 2017;3:24. [Crossref] [PubMed]
- Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133. [Crossref] [PubMed]


