
A comprehensive characterization of the transcriptome in enzalutamide resistance prostate cancer
Introduction
In 2018, there were 1,276,106 newly diagnosed cases of prostate cancer (PCa) and 358,989 associated deaths, making it the second common malignancy and the fifth leading cause of cancer-related deaths in men worldwide (1). The progression and proliferation of PCa is facilitated by the androgen-signaling axis (2,3) and thus, the main treatment for PCa involves the suppression of androgens (4,5) via anti-androgen deprivation therapy (ADT) (6). However, ADT inevitable leads to the emergence of castration-resistant prostate cancer (CRPC) (7,8).
Enzalutamide, which interferes with the androgen receptor (AR), and abiraterone, which inhibits androgen synthesis, are commonly used for the treatment of PCa. There is increasing evidence to suggest that enzalutamide may have clinical significance in the overall survival (OS) of CRPC patients with metastasis [hazard ratio (HR) =0.63; 95% confidence interval (CI): 0.53 to 0.75; P<0.001] (9) and in metastasis-free survival in patients without metastasis (HR =0.29; 95% CI: 0.24 to 0.35; P<0.001) (10). Indeed, enzalutamide has been approved for both chemotherapy-naïve and chemotherapy-exposed CRPC patients (6,9,11). However, almost all patients of CRPC eventually develops resistance to enzalutamide through complex mechanisms (8,11). Therefore, further understanding the mechanisms by which PCa develops resistance to enzalutamide is urgently required to develop novel treatments for patients with PCa.
Immune “cold tumor” mainly refers to the less infiltration of local immune cells in the tumor and the low expression of PD-L1. If there are ways to improve the tumor immune microenvironment, such as strengthening the role of immune cells, increasing immune cell infiltration, and enhancing immune antigen presentation. Further, it can increase the response rate of PCa to immunotherapy. However, enzalutamide resistance appears to be involved in immune cell infiltration. The current study examined the genomic characteristics of enzalutamide-resistant PCa. Differential gene expression analyses and protein-protein interaction (PPI) network analyses were performed to determine key enzalutamide-related genes. Univariate Cox analysis was used to identify the hub genes, contactin 2 (CNTN2) and frizzled class receptor 2 (FZD2), with prognostic implications. Further validation using the Gene Expression Omnibus (GEO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (12) database suggested that both hub genes were correlated with the AR and enzalutamide. Gene set enrichment analysis (GSEA) demonstrated that both hub genes were positively correlated to immune-related pathways. CIBERSORT (13) was used to confirm the association between the hub genes and immune cell infiltration levels. These hub genes, CNTN2 and FZD2, may be potential biomarkers or immunotherapeutic drug targets for the management of enzalutamide resistance in PCa patients.
We present the following article in accordance with the STREGA reporting checklist (available at https://dx.doi.org/10.21037/atm-21-6191).
Methods
Data source and preprocessing
The RNA-sequencing data of enzalutamide-resistant or sensitive PCa cells was derived from the GSE163240 (14) and GSE136129 (15) datasets in the GEO (16) database. The GSE147976 (17) and GSE150807 (18,19) datasets with gene expression data from enzalutamide-naïve or -exposed cells were also downloaded for validation.
Patient survival-related data were obtained from The Cancer Genome Atlas (TCGA) (20), the International Cancer Genome Consortium (ICGC) (21), cBioPortal (22,23), and the GEO database. Based on the criteria that datasets must have at least 20 samples with complete follow-up data and corresponding gene expression data to implement prognostic analysis, a total of 6 datasets were finally included. The gene expression data including fragments per kilobase of transcript per million (FPKM) values and clinical data from the prostate adenocarcinoma (PRAD) cohort in TCGA (n=493) (20) were downloaded using the R package TCGA biolinks (24,25). Survival data and gene expression data were collated from the PRAD-CA (n=144) and PRAD-FR (n=25) datasets in the ICGC database (21), the MCTP (n=35) (26) and SU2C (n=71) (27) datasets in the cBioPortal database (22,23), and the GSE116918 (n=248) (28) dataset in the GEO.
The raw counts data of PCa and normal control samples were downloaded from TCGA and Genotype-Tissue Expression (GTEx) (29) data cohorts. In addition, RNA expression data related to PCa or control samples were also derived from the GSE80609, GSE56829, and GSE111177 datasets (30). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Enzalutamide-related genes
The DESeq2 software was used to identify the differentially expressed genes between enzalutamide-resistant cells and enzalutamide-sensitive cells with a threshold of |log2FoldChange| >1 and adjusted P value <0.01. Volcano plots generated by ggplot2 (31) were used to depict these differentially expressed genes. A Venn diagram was used to intersect the differentially expressed genes from the GSE163240 and GSE136129 datasets using the ggvenn package (32). To explore the biological process (BP) involved, GSEA was used to implement functional enrichment analysis with reference to Gene Ontology (GO) (33,34), KEGG, and Molecular Signatures Database (MSigDB) (35) using the clusterProfiler (36) package.
PPI network analysis
The STRING database was used to explore the interaction among enzalutamide-related genes. A PPI network was constructed and analyzed using Cytoscape version 3.8.2 (37). Network module analysis and degree analysis were conducted to identify the key nodes using Minimal Common Oncology Data Elements (MCODE) (38) and cytoHubba (39), respectively. The key genes were derived by overlapping nodes in modules from MCODE and nodes with degree >3 from cytoHubba.
Cox analysis and determination of hub genes
Hub genes were defined as genes with prognostic implications in at least two datasets among six cohorts, including TCGA-PRAD, ICGC PRAD-CA and PRAD-FR, cBioPortal MCTP and SU2C, and GSE116918. Univariate Cox analysis was used to determine the prognostic implications with reference to OS, progression-free interval (PFI), or disease-free survival (DFS) (40). OS is the survival time from the date of diagnosis to the date of death by any cause. PFI is defined as the interval from diagnosis to the first emergence of a new tumor event (40). DFS is defined as the interval from a patient’s disease-free status after their first diagnosis and therapy to the first emergence of a new tumor event (40). A P value <0.05 is considered statistically significant.
Exploration of the biological functions of the hub genes
GSEA was applied to examine the BPs in the GO database, and the functional pathways in the KEGG database. The threshold recommended by GSEA (41) includes a minimum gene set size of 15, a maximum gene set size of 500, and an adjusted P value <0.25. The association with hallmark gene sets (35) was also determined. In addition, PCa-special scores [AR scores and neuroendocrine prostate cancer (NEPC) scores] (27,42,43) were analyzed based on the SU2C dataset. The Wilcoxon rank sum and Kruskal-Wallis rank sum tests were used to explore the expression levels of hub genes at various stages of PCa and in normal control samples.
Validation of the association with between the hub genes and enzalutamide
A differential gene expression analysis was conducted in enzalutamide-exposed PCa cells and enzalutamide-naïve PCa cells from the GSE147976 and GSE150807 datasets. Moreover, the enzalutamide-targeted functional pathways were examined using the KEGG database and GSEA was conducted to determine whether the hub genes were enriched in enzalutamide-targeted functional pathways with a threshold of P value <0.05.
Exploration of the association between the PCa hub genes and immune cell infiltration
The immune subtypes related to PCa and the genes that code immunomodulators and chemokines were identified using studies by Thorsson et al. (44) and Charoentong et al. (45), respectively. A total of 4 immune subtypes, including C1 (wound healing), C2 [interferon-gamma (IFNg) dominant], C3 (inflammatory), and C4 (lymphocyte depleted), were identified. The Kruskal-Wallis rank sum test was used to assess the association between immune subtypes and the hub genes. The Spearman correlation coefficient was calculated to explore the association between hub genes and immunomodulators or chemokines. CIBERSORT in the R program was used to enumerate the absolute infiltration levels of 22 types of immune cells in PCa. The differential infiltration immune cells between PCa samples and normal samples were determined by the Wilcoxon rank sum test. Spearman correlation coefficient analysis was performed to investigate the association between immune infiltration levels and hub genes. A P value <0.05 was considered statistically significant.
Statistical analysis
All statistical tests were based on a significant P value <0.05. The Benjamini-Hochberg method was used to adjust the P value. An adjusted P value <0.05 was applied when analyses involved multiple comparisons. The R program (version 4.0.2) (46) was used for all statistical analyses.
A schematic diagram depicting the processes involved in this study is shown in Figure 1.
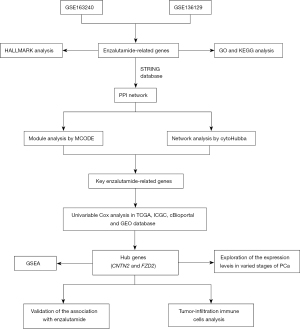
Results
Identification of the enzalutamide-related genes
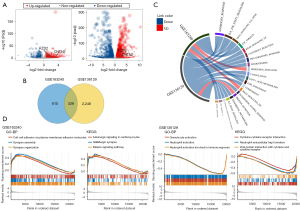
Analysis of the GSE163240 dataset comparing enzalutamide-resistant cells and enzalutamide-sensitive cells revealed a total of 686 up-regulated genes and 253 down-regulated genes (Figure 2A). In the GSE136129 dataset, there were 1,246 up-regulated genes and 1,329 down-regulated genes (Figure 2A). A Venn diagram revealed 329 overlapping enzalutamide-related genes identified in both datasets (Figure 2B). A chord diagram constructed using the circlize package (47) showed that the enzalutamide-related genes were significantly negatively correlated with androgen response, L6/JAK/STAT3 signaling, and tumor necrosis factor (TNF)α signaling via nuclear factor (NF) κB (Figure 2C). GO analysis suggested that the differentially expressed enzalutamide-related genes identified in the GSE163240 and GSE136129 datasets were enriched in BPs including cell-cell adhesion via plasma-membrane adhesion molecules, synapse assembly and organization, and granulocyte and neutrophil activation associated with the immune response (Figure 2D). KEGG analysis demonstrated that the differentially expressed enzalutamide-related were mostly associated with pathways including adrenergic signaling in cardiomyocytes, GABAergic synapse and relaxin signaling pathway, cytokine-cytokine receptor interaction, neutrophil extracellular trap formation, and viral protein interaction with cytokines and cytokine receptors (Figure 2D).
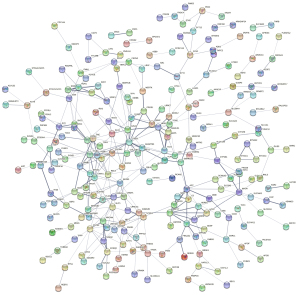
Development of the PPI network
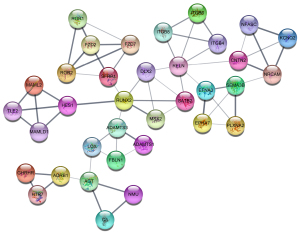
A total of 329 enzalutamide-related genes were imported into the STRING database. A network including 321 nodes and 355 edges was constructed (Figure 3), with an enrichment P value of 1.0e-16, suggesting that this network has significantly more interactions than expected. Furthermore, module analysis using MCODE extracted 7 clusters, including 53 genes. The largest module is depicted in Figure 4. In addition, cytoHubba analysis suggested that there are 70 genes with a degree >3. By overlapping these genes, 37 key genes were identified for further analysis.
Determination of hub genes
To obtain genes with prognostic significance to PCa, univariate Cox analysis was applied in the TCGA, PRAD-CA, PRAD-FR, MCTP, SU2C, and GSE116918 datasets.
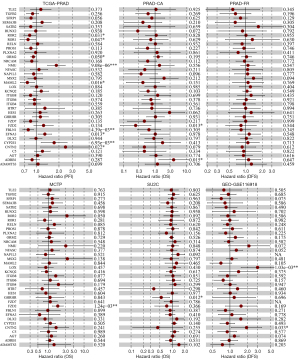
The TCGA dataset identified 9 genes that were significantly correlated with PFI, including CNTN2 (HR =1.140–5.225; P<0.05), EFNA3 (HR =1.058–1.602; P<0.05), ROR2 (HR =0.686–0.983; P<0.05), FBLN1 (HR =0.665–0.927; P<0.05), NMU (HR =1.408–2.421; P<0.05), MAML2 (HR =0.615–0.950; P<0.05), ROR1 (HR =0.611–0.997; P<0.05), ORM2 (HR =0.842–1.000; P<0.05), and CYP2E1 (HR =1.162–2.567; P<0.05). Analysis of the PRAD-CA dataset revealed that the genes ADRB1 (HR =1.308–11.696; P=0.015) and FZD2 (HR =0.078–0.743; P=0.013) were significantly correlated with OS. The MCTP and SU2C datasets, respectively, demonstrated that FZD2 (HR =1.264–2.603; P=<0.05) and GHRHR (HR =0.335–0.874; P<0.05) were correlated with OS. The GSE116918 dataset further suggested that both CNTN2 (HR =1.053–3.529; P<0.05) and LOX (HR =1.553–5.727; P<0.05) significantly impacted DFS in PCa patients. However, examination of the PRAD-FR dataset revealed that there was no significant correlation among any of the genes (Figure 5). Therefore, the CNTN2 and FZD2 genes were considered hub genes due to their significant implication in PCa prognosis in at least two datasets.
Biological function of the hub genes
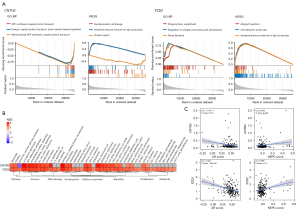
GSEA was used to depict the biological function of the hub genes. As shown in Figure 6A, CNTN2 was strongly correlated with BPs including ATP synthesis coupled proton transport, energy coupled proton transport, down electrochemical gradient, and mitochondrial ATP synthesis coupled proton transport. FZD2 was associated with BPs involving phagocytosis, engulfment, regulation of antigen processing and presentation, and renal filtration. KEGG analysis suggested that both CNTN2 and FZD2 were associated with the intestinal immune network for immunoglobulin (Ig)A production (Figure 6A). Hallmark gene sets revealed that both hub genes were highly associated with immune-related pathways (such as allograft rejection, inflammatory response, IFNg response, IFNα response, IL6/JAK/STAT3 signaling, and complement and coagulation) and proliferation-related pathways (such as mitotic spindle and the p53 pathway) (Figure 6B). Analysis of the SU2C dataset reveal that both hub genes were negatively associated with AR scores [correlation coefficient (R) =−0.172 and P<0.05 for CNTN2; and R=−0.327 and P<0.05 for FZD2; Figure 6C]. Furthermore, FZD2 was positively correlated to NEPC scores (R=0.176 and P<0.05; Figure 6C).
The importance of the hub genes in PCa
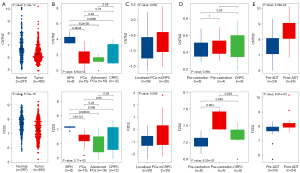
Both CNTN2 and FZD2 hub genes were significantly down-regulated in PCa samples compared to normal healthy tissues obtained from the TCGA and GTEx data cohorts (P<0.05 for CNTN2; P<0.05 for FZD2; Figure 7A). The GSE80609 dataset also verified the significant differential expression of the hub genes in both low-grade PCa (P=0.00034 and 1.9e-05 for CNTN2 and FZD2, respectively) and advanced PCa (P=8.2e-05 and 0.00033 for CNTN2 and FZD2, respectively) compared to benign prostatic hyperplasia (BPH). In addition, CNTN2 was also significantly differentially expressed in CRPC (P=0.0096; Figure 7B). However, in the GSE80609 dataset, there was no significant difference in the expression of either hub genes across different stages of PCa (Figure 7B). The MCTP dataset comparing metastatic CRPC (mCRPC) with localized PCa showed statistical difference in the expression of FZD2 (P=0.029; Figure 7C). Furthermore, the correlation between hub gene expression and castration was examined. The expression of FZD2 was up-regulated after castration (P=8.32e-03; Figure 7D) and ADT (P=6.91e-03; Figure 7E). However, in the GSE56829 dataset, there was no significant difference in CNTN2 expression after castration (P=0.894; Figure 7D), while the GSE111177 dataset indicated that CNTN2 expression was significantly associated with ADT (P=3.59e-04; Figure 7E).
Validation of the association between the hub genes and enzalutamide
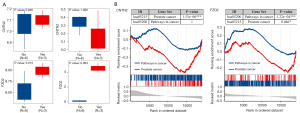
Differential expression analysis in the GSE147976 and GSE150807 datasets showed that FZD2 was significant up-regulated in the enzalutamide-exposed group compared to the enzalutamide-naïve group, while there were no statistical differences in CNTN2 expression (Figure 8A). We got both gene sets targeted by enzalutamide after searching KEGG database (hsa05215: PCa and hsa05200: pathways in cancer) (48). GSEA suggested that both hub genes were enriched in PCa pathways (P<0.05 both for CNTN2 and FZD2) and FZD2 was also related to cancer pathways (P=1.22e-04; Figure 8B).
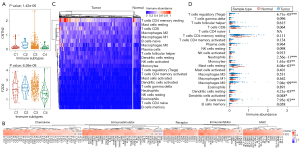
Analysis of the immune infiltration in PCa
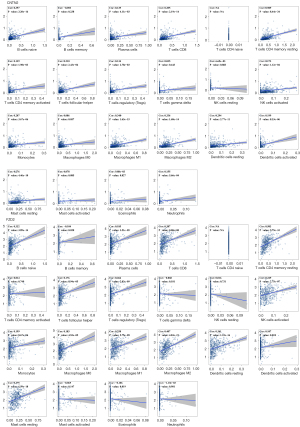
The Kruskal-Wallis rank sum test indicated that both hub genes were differentially expressed across different immune subtypes (P=1.42e-06 and 6.58e-08 for CNTN2 and FZD2, respectively; Figure 9A). Interestingly, both hub genes showed positive correlation with most immunomodulators, chemokines, receptors, and major histocompatibility complexes (MHCs) (Figure 9B). CIBERSORT was applied to estimate the infiltration abundance of 22 types of immune cells in PCa and normal control samples (Figure 9C). There was a significant difference in the level of infiltration of regulatory T cells (P=6.71e-05), neutrophils (P=2.56e-11), monocytes (P=1.61e-03), resting mast cells (P=6.04e-03), M0 macrophages (P=7.04e-09), resting dendritic cells (P=4.21e-03), activated dendritic cells (P=0.048), and naïve B cells (P=7.95e-03) between PCa and normal control samples (Figure 9D). Spearman correlation analysis indicated that both hub genes exhibited positive correlation with the infiltration abundance of most of immune cells (resting mast cells, activated dendritic cells, resting dendritic cells, M2 macrophages, M1 macrophages, monocytes, activated natural killer (NK) cells, regulatory T cells, follicular helper T cells, CD4 memory resting T cells, CD8 T cells, plasma cells, and naïve B cells) (Figure 10).
Discussion
PCa is the most common cancer and the second leading cause of cancer-related deaths in men in United States. In 2020, there were an estimated 191,930 new cases and 33,330 related deaths (49). Unfortunately, resistance to enzalutamide presents a significant challenge in the treatment of PCa. The present study explored the underlying molecular characteristics of enzalutamide-resistant PCa using data obtained from the GEO, ICGC, cBioPortal, TCGA, and GTEx databases.
Enzalutamide-related genes were determined by differential expression analysis. As expected, these genes were highly inversely correlated with androgen response in both gene datasets (GSE163240 and GSE136129) (Figure 2C). Previous studies have reported that resistance to enzalutamide is associated with the AR signaling pathway (8,11). In addition, the enrichment of the IL6/JAK/STAT3 signaling pathway in both gene sets indicated that the resistance of enzalutamide may be related to immune-associated functions in PCa. GO and KEGG analysis verified that enzalutamide resistance is related to immune activity (including cell-cell adhesion via plasma-membrane adhesion molecules, granulocyte activation, and neutrophil activation), and this is consistent with previous reports (50,51).
Network analysis and univariate Cox analysis identified two hub genes, CNTN2 and FZD2, both of which were significantly correlated with the AR. The expression of CNTN2 and FZD2 were significantly different at various stages of PCa compared to normal healthy samples. Furthermore, both hub genes were significantly associated with ADT. Interestingly, the expression of FZD2 was significantly up-regulated in enzalutamide-exposed samples, while no statistical difference was found for CNTN2, and this may have been due to the small sample size. These findings suggested that both hub genes, CNTN2 and FZD2, play a vital role in the resistance of PCa cells to enzalutamide.
The GSEA results indicated that both hub genes were highly correlated with immune-associated functional pathways, including allograft rejection, inflammatory response, IFNg response, IFNα response, IL6/JAK/STAT3 signaling, and complement and coagulation. Furthermore, both hub genes showed significantly positive correlations with the expression of almost all immunomodulators, chemokines, receptors, MHCs, and the infiltration of 22 types of immune cells. This suggested that the resistance of enzalutamide may be related to immune infiltration (50,51) and thus, both hub genes may be potential immunotherapeutic targets in PCa.
To date, there have been limit studies relating to CNTN2 and most research have been conducted in gliomas. CNTN2 is highly up-regulated in oligodendrogliomas compared with glioblastomas (52). It has also been reported to coamplify with MDM4, which is involved in the progression of malignant gliomas (53). In addition, CNTN2 has been shown to mediate the RTK/RAS/MAPK pathway in glioma cells (54). Although there is a paucity of data regarding CNTN2 and PCa, studies have suggested that the RAS/MAPK pathway may be involved in the development and progression of PCa and in the resistance to castration (55-57). Indeed, the RAS/MAPK pathway interacting with PI3K-AKT-mTOR has been implicated in many drug-resistance mechanisms including enzalutamide, ipatasertib, and capivasertib (57-59). Therefore, it is possible that CNTN2 may play an important role in mediating enzalutamide resistance in PCa cells.
FZD2, which is a receptor for Wnts, has been implicated in both non-canonical and canonical Wnt signaling pathways in PCa (8,60). FZD2 can regulate the epithelial-mesenchymal transition (EMT) pathway which is characterized by promoting tissue differentiation and shaping (61), and is correlated with the proliferation and metastasis of breast, endometrial, colon, liver, lung, and salivary adenoid cystic carcinomas (62-68). There is strong evidence that the EMT pathway is related to the metastasis and development of NEPC and CRPC (69-71). In addition, the activation of Wnt signaling pathway is one of the mechanisms leading to enzalutamide resistance in PCa cells (8). All these studies suggested that FZD2 may be an effective biomarker for enzalutamide-resistant PCa.
This current study comprehensively explored potential enzalutamide-related biomarkers in PCa. However, there were some limitations to this investigation. Further in vitro and in vivo studies should be conducted to further examine the detailed molecular mechanisms by which CNTN2 and FZD2 mediate enzalutamide-resistance in PCa cells.
In conclusion, this study analyzed the genetic characteristics of enzalutamide-resistant PCa and identified two hub genes, namely, CNTN2 and FZD2, that are related to immune infiltration in PCa. These findings provide crucial information regarding the mechanisms of enzalutamide-resistance in PCa. These hub genes may serve as potential biomarkers or immunotherapeutic drug targets for the management of enzalutamide resistance in PCa.
Acknowledgments
The authors would like to acknowledge the TCGA database (https://portal.gdc.cancer.gov/), the GTEx database (https://www.gtexportal.org/), the GEO database (https://www.ncbi.nlm.nih.gov/geo/), the ICGC database (https://dcc.icgc.org/), and the cBioPortal database (https://www.cbioportal.org/) for provision of data.
Funding: This work was supported by the National Natural Science Foundation of China (Program No. 81572543) and the Science and Technology Support Program of Tianjin, China (Grant No. 17ZXMFSY00040).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-6191
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-6191). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Dai C, Heemers H, Sharifi N. Androgen Signaling in Prostate Cancer. Cold Spring Harb Perspect Med 2017;7:a030452. [Crossref] [PubMed]
- Mohler JL. Castration-recurrent prostate cancer is not androgen-independent. Adv Exp Med Biol 2008;617:223-34. [Crossref] [PubMed]
- Mills IG. Maintaining and reprogramming genomic androgen receptor activity in prostate cancer. Nat Rev Cancer 2014;14:187-98. [Crossref] [PubMed]
- Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer 2015;15:701-11. [Crossref] [PubMed]
- Litwin MS, Tan HJ. The Diagnosis and Treatment of Prostate Cancer: A Review. JAMA 2017;317:2532-42. [Crossref] [PubMed]
- Guo F, Li GH. Enzalutamide alleviates anxiety and depression as well as improves quality of life compared to bicalutamide in metastatic castration-resistant prostate cancer patients: a cohort study. Transl Cancer Res 2019;8:1965-74. [Crossref]
- Wang Y, Chen J, Wu Z, et al. Mechanisms of enzalutamide resistance in castration-resistant prostate cancer and therapeutic strategies to overcome it. Br J Pharmacol 2021;178:239-61. [Crossref] [PubMed]
- Sartor O, de Bono JS. Metastatic Prostate Cancer. N Engl J Med 2018;378:645-57. [Crossref] [PubMed]
- Hussain M, Fizazi K, Saad F, et al. Enzalutamide in Men with Nonmetastatic, Castration-Resistant Prostate Cancer. N Engl J Med 2018;378:2465-74. [Crossref] [PubMed]
- Tucci M, Zichi C, Buttigliero C, et al. Enzalutamide-resistant castration-resistant prostate cancer: challenges and solutions. Onco Targets Ther 2018;11:7353-68. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Chen B, Khodadoust MS, Liu CL, et al. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol 2018;1711:243-59. [Crossref] [PubMed]
- Furlan T, Kirchmair A, Sampson N, et al. MYC-Mediated Ribosomal Gene Expression Sensitizes Enzalutamide-resistant Prostate Cancer Cells to EP300/CREBBP Inhibitors. Am J Pathol 2021;191:1094-107. [Crossref] [PubMed]
- He Y, Wei T, Ye Z, et al. A noncanonical AR addiction drives enzalutamide resistance in prostate cancer. Nat Commun 2021;12:1521. [Crossref] [PubMed]
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 2013;41:D991-5. [Crossref] [PubMed]
- Zhang Z, Karthaus WR, Lee YS, et al. Tumor Microenvironment-Derived NRG1 Promotes Antiandrogen Resistance in Prostate Cancer. Cancer Cell 2020;38:279-96.e9. [Crossref] [PubMed]
- Verma S, Shankar E, Chan ER, et al. Metabolic Reprogramming and Predominance of Solute Carrier Genes during Acquired Enzalutamide Resistance in Prostate Cancer. Cells 2020;9:2535. [Crossref] [PubMed]
- Verma S, Shankar E, Kalayci FNC, et al. Androgen Deprivation Induces Transcriptional Reprogramming in Prostate Cancer Cells to Develop Stem Cell-Like Characteristics. Int J Mol Sci 2020;21:9568. [Crossref] [PubMed]
- GDC TCGA portal. [cited 2020 Mar 27]. Available online: https://portal.gdc.cancer.gov/
- ICGC Data Portal. [cited 2021 Jan 7]. Available online: https://dcc.icgc.org/
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016;44:e71. [Crossref] [PubMed]
- Mounir M, Lucchetta M, Silva TC, et al. New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput Biol 2019;15:e1006701. [Crossref] [PubMed]
- Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487:239-43. [Crossref] [PubMed]
- Abida W, Cyrta J, Heller G, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019;116:11428-36. [Crossref] [PubMed]
- Jain S, Lyons CA, Walker SM, et al. Validation of a Metastatic Assay using biopsies to improve risk stratification in patients with prostate cancer treated with radical radiation therapy. Ann Oncol 2018;29:215-22. [Crossref] [PubMed]
- GTEx Portal. [cited 2020 Mar 27]. Available online: https://www.gtexportal.org/home/
- Sharma NV, Pellegrini KL, Ouellet V, et al. Identification of the Transcription Factor Relationships Associated with Androgen Deprivation Therapy Response and Metastatic Progression in Prostate Cancer. Cancers (Basel) 2018;10:379. [Crossref] [PubMed]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag, 2016. Available online: https://ggplot2.tidyverse.org
- Yan L. ggvenn: Draw Venn Diagram by “ggplot2”. 2021. Available online: https://cran.uvigo.es/web/packages/ggvenn/ggvenn.pdf
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 2019;47:D330-8. [Crossref] [PubMed]
- Liberzon A, Birger C, Thorvaldsdóttir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417-25. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. [Crossref] [PubMed]
- Liu J, Lichtenberg T, Hoadley KA, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173:400-16.e11. [Crossref] [PubMed]
- GSEA User Guide. [cited 2021 Jan 8]. Available online: https://www.gsea-msigdb.org/gsea/doc/GSEAUserGuideFrame.html
- Beltran H, Prandi D, Mosquera JM, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016;22:298-305. [Crossref] [PubMed]
- Hieronymus H, Lamb J, Ross KN, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell 2006;10:321-30. [Crossref] [PubMed]
- Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity 2018;48:812-30.e14. [Crossref] [PubMed]
- Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 2017;18:248-62. [Crossref] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing, 2020. Available online: https://www.R-project.org/
- Gu Z, Gu L, Eils R, et al. circlize Implements and enhances circular visualization in R. Bioinformatics 2014;30:2811-2. [Crossref] [PubMed]
- KEGG DRUG: Enzalutamide. [cited 2021 Apr 16]. Available online: https://www.genome.jp/dbget-bin/www_bget?dr:D10218
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Ruan H, Bao L, Tao Z, et al. Flightless I Homolog Reverses Enzalutamide Resistance through PD-L1-Mediated Immune Evasion in Prostate Cancer. Cancer Immunol Res 2021;9:838-52. [Crossref] [PubMed]
- Madan RA, Karzai F, Donahue RN, et al. Clinical and immunologic impact of short-course enzalutamide alone and with immunotherapy in non-metastatic castration sensitive prostate cancer. J Immunother Cancer 2021;9:e001556. [Crossref] [PubMed]
- Yu F, Fu WM. Identification of differential splicing genes in gliomas using exon expression profiling. Mol Med Rep 2015;11:843-50. [Crossref] [PubMed]
- Riemenschneider MJ, Knobbe CB, Reifenberger G. Refined mapping of 1q32 amplicons in malignant gliomas confirms MDM4 as the main amplification target. Int J Cancer 2003;104:752-7. [Crossref] [PubMed]
- Yan Y, Jiang Y. RACK1 affects glioma cell growth and differentiation through the CNTN2-mediated RTK/Ras/MAPK pathway. Int J Mol Med 2016;37:251-7. [Crossref] [PubMed]
- Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18:11-22. [Crossref] [PubMed]
- Armenia J, Wankowicz SAM, Liu D, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet 2018;50:645-51. [Crossref] [PubMed]
- Shorning BY, Dass MS, Smalley MJ, et al. The PI3K-AKT-mTOR Pathway and Prostate Cancer: At the Crossroads of AR, MAPK, and WNT Signaling. Int J Mol Sci 2020;21:4507. [Crossref] [PubMed]
- Adelaiye-Ogala R, Gryder BE, Nguyen YTM, et al. Targeting the PI3K/AKT Pathway Overcomes Enzalutamide Resistance by Inhibiting Induction of the Glucocorticoid Receptor. Mol Cancer Ther 2020;19:1436-47. [Crossref] [PubMed]
- Bitting RL, Armstrong AJ. Targeting the PI3K/Akt/mTOR pathway in castration-resistant prostate cancer. Endocr Relat Cancer 2013;20:R83-99. [Crossref] [PubMed]
- Zins K, Schäfer R, Paulus P, et al. Frizzled2 signaling regulates growth of high-risk neuroblastomas by interfering with β-catenin-dependent and β-catenin-independent signaling pathways. Oncotarget 2016;7:46187-202. [Crossref] [PubMed]
- Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871-90. [Crossref] [PubMed]
- Gujral TS, Chan M, Peshkin L, et al. A noncanonical Frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell 2014;159:844-56. [Crossref] [PubMed]
- Huang L, Luo EL, Xie J, et al. FZD2 regulates cell proliferation and invasion in tongue squamous cell carcinoma. Int J Biol Sci 2019;15:2330-9. [Crossref] [PubMed]
- Yin P, Wang W, Gao J, et al. Fzd2 Contributes to Breast Cancer Cell Mesenchymal-Like Stemness and Drug Resistance. Oncol Res 2020;28:273-84. [Crossref] [PubMed]
- Ding LC, Huang XY, Zheng FF, et al. FZD2 inhibits the cell growth and migration of salivary adenoid cystic carcinomas. Oncol Rep 2016;35:1006-12. [Crossref] [PubMed]
- Saleh AD, Cheng H, Martin SE, et al. Integrated Genomic and Functional microRNA Analysis Identifies miR-30-5p as a Tumor Suppressor and Potential Therapeutic Nanomedicine in Head and Neck Cancer. Clin Cancer Res 2019;25:2860-73. [Crossref] [PubMed]
- Asano T, Yamada S, Fuchs BC, et al. Clinical implication of Frizzled 2 expression and its association with epithelial-to-mesenchymal transition in hepatocellular carcinoma. Int J Oncol 2017;50:1647-54. [Crossref] [PubMed]
- Bian Y, Chang X, Liao Y, et al. Promotion of epithelial-mesenchymal transition by Frizzled2 is involved in the metastasis of endometrial cancer. Oncol Rep 2016;36:803-10. [Crossref] [PubMed]
- Grant CM, Kyprianou N. Epithelial mesenchymal transition (EMT) in prostate growth and tumor progression. Transl Androl Urol 2013;2:202-11. [PubMed]
- Nouri M, Ratther E, Stylianou N, et al. Androgen-targeted therapy-induced epithelial mesenchymal plasticity and neuroendocrine transdifferentiation in prostate cancer: an opportunity for intervention. Front Oncol 2014;4:370. [Crossref] [PubMed]
- Chen R, Dong X, Gleave M. Molecular model for neuroendocrine prostate cancer progression. BJU Int 2018;122:560-70. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


