
Regulatory mechanism of HIF-1α and its role in liver diseases: a narrative review
Introduction
The liver is not only the most important organ for metabolism and detoxification but also the main site of many biological processes (1-4). Indeed, the liver plays a very important role in metabolism (5), bile production (6), detoxification (7), blood coagulation (8), immunity (9), and heat generation as well as the regulation of water and electrolyte contents (10). Many factors negatively affect the liver, such as lack of sleep, alcohol consumption, gastrointestinal bleeding, infection, portal vein thrombosis, dehydration, and kidney failure (11,12). Overall, liver disease, which is increasing in incidence and severity, is a serious health problem, resulting in considerable economic and social burdens (13-15).
In the early 1990s, Semenza et al. (16) discovered HIF while studying EPO gene expression. The 2019 Nobel Prize in Physiology or Medicine was awarded for work related to how cells detect and adapt to different oxygen environments, with one of the most critical factors being hypoxia-inducible factor (HIF), a heterodimeric transcription factor that consists of α and β subunits. Expression of the α subunit depends on oxygen, whereas the β subunit is constitutively expressed (17-19). HIF-1α is the main regulator of hypoxia signaling and is widely expressed (20-22).
HIF-1α is involved in the occurrence and development of various liver diseases, including alcoholic liver disease (ALD), nonalcoholic fatty liver disease (NAFLD), viral hepatitis, liver fibrosis, drug-induced liver injury and hepatocellular carcinoma (HCC) (23,24). Although HIF-1α is expected to become a new target for the treatment of liver diseases, the development of strategies involving HIF-1α regulation remains a challenge. This review discusses and summarizes the role and potential mechanism of action of HIF-1α in some common liver diseases to provide new methods and ideas for their treatment.
We present the following article in accordance with the Narrative Review reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-4222/rc).
The characteristics of HIF-1α
Structure of HIF-1α
As previously described, HIF proteins constitute a family of transcriptional regulators that play a central role in regulating gene expression under conditions of low oxygen (25,26). The HIF family is composed of HIF-1α, HIF-2α, and HIF-3α, each of which dimerizes with HIF-1β, also known as the aryl-hydrocarbon-nuclear receptor translocator (ARNT), and less frequently with ARNT2 (27). HIFs belong to the subfamily of PER-ARNT-SIM (PAS) transcription factors in the basic helix-loop-helix (bHLH) family (28). The structures of the α subunit and β subunit are similar, and both contain the following domains: an N-terminal bHLH domain, which binds to DNA; a middle region PAS domain, which promotes heterodimer formation; and a C-terminal domain, which binds with transcriptional cofactors to promote transcriptional coregulation (29,30). The most common HIF protein is HIF-1α, which has the most wide-ranging effects (31).
Degradation and activation of HIF-1α
The state of the α subunit determines the stability of HIF-1. The α subunit is degraded rapidly under normoxic conditions but is stable during hypoxia (32-34). Hydroxylation of proline residues by proline hydroxylase domain enzymes (PHDs) is a key step in the degradation of the α-subunit (32,35,36): under normoxia, the conserved proline residues 402 and 564 are hydroxylated by prolyl hydroxylases (PHD1, PHD2, or PHD3) (37). Subsequently, von Hippel Lindau protein (pVHL), a tumor-suppressor E3 ubiquitin ligase component, mediates ubiquitination of HIF-1α through specific binding to the two hydroxylated proline residues, promoting rapid degradation of the α subunit via the ubiquitin proteasome pathway (38,39). Factor-inhibiting HIF-1 (FIH1) and reactive oxygen species (ROS) can also affect HIF-1α stability. The former hydroxylates an asparagine residue (Asn803) of HIF-1α in the C-terminal transactivation domain, thereby blocking HIF binding to the transcriptional coactivator cAMP response element-binding protein (CBP)/p300 and inhibiting transcriptional activation of HIF-1α (40-43). FIH1 also interacts with pVHL to serve as a coinhibitor to suppress transactivation by recruiting histone deacetylases (HDACs) (43). ROS prevent HIF-1α degradation by blocking PHD activation to inhibit acetylation of HIF-1α (44). Acetylation has a profound and complex effect on the stability of the HIF-1α protein. For example, Seo et al. (45) found that HDAC4 and HDAC5 enhance the transactivation function of HIF-1 by promoting dissociation of HIF-1 from FIH-1 and association with p300. In addition, Kang et al. (46) reported that FIH-1 hydroxylates hARD1/NAA10, a component of N-terminal acetyltransferase (NatA), under normoxia and thus promotes pVHL binding to HIF-1α via acetylation. There are also reports that HIF1α is stabilized by p300 via Lys-709 acetylation (47), and SIRT2-mediated HIF-1α deacetylation is critical for destabilization of HIF-1α (48). PHD activity is restricted by oxygen availability (41). In the absence of oxygen, the hydroxylation of HIF-1α is also inhibited, causing the HIF-1α subunit to become stable and accumulate in the cytoplasm (49). The accumulated HIF-1α is then transferred to the nucleus, forms dimers with HIF-1β (ARNT), and interacts with the transcriptional coactivator CBP/p300 to form a transcription initiation complex that recognizes hypoxia response elements (HREs) in the promoters of target genes to induce transcription (23,39,44,50) (Figure 1).
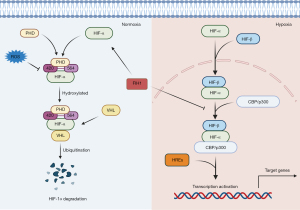
In addition, the stability of HIF-1α is affected by some factors under nonhypoxic conditions, including metals, growth factors, pH, and mechanical stress (51,52).
Genes targeted by HIF-1α
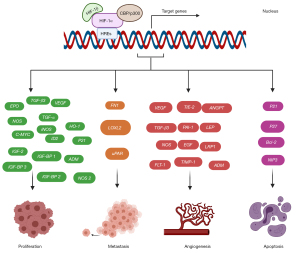
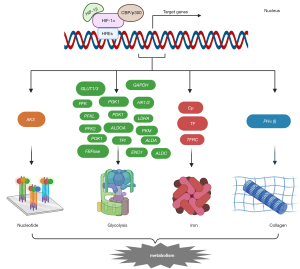
As mentioned above, HIF-1α is transferred to the nucleus under hypoxic conditions, where it recognizes HREs in the promoters of target genes to promote transcription. In fact, HIF-1α induces the transcription of many genes (currently over 2,000 suggested genes and more than 300 known genes) and is widely involved in various biological processes, including the following: tumor-related cell proliferation, metastasis, angiogenesis and apoptosis (Figure 2); metabolism-related glycolysis metabolism, nucleotide metabolism, iron metabolism and collagen metabolism (Figure 3); inflammation and immunity; erythropoiesis; pH regulation; and others (51-55). This review focuses on most of the genes involved in the above processes for the convenience of future research on pathways downstream of HIF-1α. Genes related to inflammation and immunity mainly include tumor necrosis factor α (TNF-α), recombination activating genes (RAGS), potassium channels in B cells (Task-2) and CD18 (56-58). Erythropoiesis is associated with erythropoietin (EPO) (59), and pH adjustment is associated with monocarboxylate transporter 4 (MCT4) (60-63) and membrane-associated carbonic anhydrase IX (CA9) (64).
HIF-1α in liver diseases
This review mainly summarizes the role and mechanism of HIF-1α in liver diseases such as ALD, NAFLD, acetaminophen (APAP)-induced liver injury (AILI), viral hepatitis, and HCC to provide a new perspective for exploring potential therapeutic targets.
ALD
Long-term alcohol intake can seriously affect liver function, leading to liver hypoxia, steatosis, and eventually ALD development (65,66). ALD is the general term for a series of diseases that range from simple liver steatosis to severe alcoholic hepatitis, followed by liver fibrosis and cirrhosis, which may eventually lead to HCC (67,68). ALD is a major cause of morbidity and mortality worldwide (69-71); its pathogenesis is complex, and there is still a lack of effective targeted treatment methods. Recent research has shown that HIF-1α is involved in the pathogenesis of ALD. However, the specific mechanism of action of HIF-1α in ALD remains unclear, a situation that is complicated by conflicting evidence. Indeed, some studies indicate that HIF-1α plays a protective role in ALD, whereas others suggest that HIF-1α aggravates liver damage and liver steatosis in ALD.
Nevertheless, a number of mechanisms support the protective effects of HIF-1α. Impaired intestinal barrier function and oxidative stress injury play a key role in the pathogenesis of ALD (72). Alcohol intake can cause intestinal barrier dysfunction, which mainly includes increased intestinal permeability, bacterial translocation, and release of endotoxin into the circulation (73). In addition, alcohol-induced inflammatory reactions and oxidative stress exacerbate tissue hypoxia (74). It is well known that long-term alcohol intake increases liver oxygen consumption, resulting in hypoxia around the center of the organ (75,76), leading to the occurrence, development, and further aggravation of ALD. Lin and colleagues (77) found that alcohol causes thinning and dysbiosis of intestinal villi, reduces expression of genes such as HIF-1α, occludin, GPX1 and SOD1, and impairs intestinal barrier function. Dietary copper supplementation can alleviate the intestinal morphology and dysfunction caused by alcohol to contribute to a cure for ALD. According to Shao et al. (78), a lack of intestinal HIF-1α can aggravate ALD by inducing intestinal disorders and barrier dysfunction. Previous studies have also suggested that treatment with Lactobacillus rhamnosus GG enhances expression of intestinal hypoxia-inducing factors, promotes intestinal integrity, and reduces alcoholic liver injury (79). Moreover, Nishiyama et al. (80) reported that hepatocyte-specific deletion of the HIF-1α gene in mice exacerbates alcoholic hepatic steatosis and causes alterations in hepatic gene expression, leading to increased fatty acid synthesis via inhibition of DEC1 induction.
The opposite view is that alcohol damage activates expression of HIF-1α mRNA (81,82), and interleukin (IL)-8 aggravates alcoholic fatty liver in mice through the Akt/HIF-1α pathway (83). Satishchandran et al. (84) found that HIF-1α mRNA levels in the liver tissues of patients and ALD mice were increased compared with the control group. MiR-122 appears to protect the liver from ethanol-induced damage by decreasing HIF-1α expression. Ethanol can also induce an increase in portal pressure, which depends on upregulation of endothelin-1 expression mediated by HIF-1α (85). These findings indicate that HIF-1α acts as a mediator of proinflammatory and vasoconstrictive phenotype development in ALD (86). Furthermore, Nath et al. (87) confirmed that the accumulation of lipids in liver cells caused by alcohol consumption involves activation of HIF-1α. Jin and colleagues (88) reported that oroxylin A reduces accumulation of lipid droplets associated with lipid metabolism regulation genes and significantly inhibits nuclear translocation of HIF-1α in ethanol-treated cells. In general, oroxylin A prevents and treats alcohol-induced liver steatosis by inhibiting HIF-1α. Moreover, vitamin C reduces the level of HIF-1α protein expression and lipid accumulation (89).
In summary, HIF-1α plays a complex but indispensable role in the occurrence and development of ALD, and targeting HIF-1α in the liver may be therapeutic for ALD.
NAFLD
NAFLD is a common chronic liver disease associated with systemic metabolic disorders (90,91). Recently, experts have begun to replace the concept of NAFLD with metabolic-associated fatty liver disease (MAFLD). The prevalence of NAFLD is 20–30% in the general population and as high as 75–100% among individuals with obesity (92). Although most patients with NAFLD have no overt clinical symptoms, 20% experience progression to chronic hepatitis, which in turn leads to cirrhosis, portal hypertension, HCC, and eventually mortality. The specific pathogenesis of NAFLD has not been fully elucidated to date, and no effective targeted therapy is yet available.
As mentioned above, HIF-1α plays a very important role in NAFLD, with a mechanism of action that is mainly divided into a protective effect by inhibiting excessive accumulation of liver fat and a damaging effect by promoting liver fibrosis. In the study by Arai et al. (90), HIF-1α-induced lipin1 expression prevented abnormal lipid accumulation by inhibiting peroxisome fatty acid oxidation, though Finck et al. (93) found that lipin1 also functions as a coactivator for PPARα and PGC-1α to promote fatty acid beta-oxidation in the liver. HIF-1 regulates lipid metabolism in a specific way in the liver by sensing the cellular microenvironment under different conditions. He et al. (94) confirmed that silencing HIF-1α aggravates NAFLD in vitro by inhibiting PPAR-α/ANGPTL4 signaling. Another study using HepG2 cells also reported a protective effect of HIF-1α expression against fatty acid-induced toxicity (95). Regardless, a large number of studies have shown that hepatocyte HIF-1 promotes hepatic fibrosis in NAFLD (96), primarily by activating the PTEN/p65 signaling pathway (97). By inhibiting Nrf2-mediated oxidative stress and inhibiting expression of a variety of fibrosis factors through the miR-122/HIF-1α signaling pathway, isochlorogenic acid B (ICAB) has a significant protective effect against fibrosis in nonalcoholic steatohepatitis (NASH) (98). In addition, HIF-2α is also an important regulator of liver lipid metabolism (99-101). HIF-2α promoted the progression of NAFLD by triggering the release of serum-rich glycoproteins from liver cells (100). During obesity, activation of intestinal HIF-2α can lead to liver cirrhosis (101).
AILI
Many factors can cause liver injury (102-106). Below, we describe the mechanism and role of HIF-1α in AILI. AILI is the most common drug-induced liver injury and the main cause of acute liver failure in Western countries (107). Studies have shown that HIF-1α participates in the early stages of APAP toxicity (108). For instance, HIF-1α-deficient mice show reduced production of thrombin and plasminogen activator inhibitor-1, indicating that HIF-1α signaling contributes to hemostasis in APAP liver toxicity. A previous study also demonstrated accumulation of neutrophils in the liver of HIF-1α-deficient mice and a decrease in plasma concentrations of IL-6 and regulated on activation, normal T cell expressed and secreted (RANTES), indicating a change in the inflammatory response (108). Suzuki et al. (109) also found that HIF-1α gene deletion in T cells aggravates the acute inflammatory response induced by APAP. The underlying mechanism involves abnormal recruitment of natural-like γδT cells, increasing excessive neutrophil infiltration in the liver. In addition, anti-PHD2 promotes angiogenesis in vivo by upregulating the protein and mRNA levels of HIF-1α target genes, significantly reducing high ALT and AST activities, and significantly improving APAP-induced lobular central necrosis (110). In summary, in the pathogenesis of APAP toxicity, HIF-1α helps to reduce bleeding, aseptic inflammation and early hepatocyte necrosis.
Viral hepatitis
Some studies have clarified the role of HIF-1α in the pathogenesis of viral hepatitis, mainly with regard to hepatitis B and C. Hepatitis B virus (HBV) encodes the viral tumor protein transactivator protein X (HBx), which promotes extracellular matrix modification through the HIF/LOX pathway in liver cancer (111), and HBx mutation affects the activation of HIF-1α in HCC to varying degrees (112). Direct interaction of HBx with the bHLH/PAS domain of HIF-1α decreases the binding of pVHL to HIF-1α and prevents ubiquitin-dependent degradation of HIF-1α. HBx can also induce angiogenesis by stabilizing HIF-1α (113). A previous study demonstrated that hepatitis C virus (HCV) infection enhances autotaxin protein expression by hypoxia-induced transcription factors and provides an environment in the liver that promotes fibrosis and liver injury (114). Furthermore, HCV-associated mitochondrial dysfunction facilitates HIF-1α-mediated glycolytic adaptation (115). The HCV glycoprotein interferes with tight junctions and adhesion connexins and promotes HCC migration and the epithelial to mesenchymal transition (EMT) by stabilizing HIF-1α (116).
HCC
According to global cancer statistics, liver cancer is the third leading cause of cancer-related death (117,118), and chronic HBV and HCV infection, smoking, excessive alcohol consumption, and aflatoxin exposure, among others, are the main causes (119-121). Among these, HBV infection is the most common inducer, especially in Asia (122). The standard treatment for liver cancer includes surgical resection, transarterial embolization, radiation therapy, and chemotherapy (123-126). However, due to delays in diagnosis and the presence of metastasis, these treatments are often inadequate, eventually leading to the development of advanced HCC (127). Moreover, many patients are at an advanced stage of HCC at the time of diagnosis. Therefore, early prevention and diagnosis of liver cancer have become the main research directions.
A large number of previous studies have found that HIF-1α is widely involved in the occurrence and development of liver cancer. HIF-1α overexpression indicates a poor prognosis for HCC patients (128,129). HIF-1α is mainly involved in promoting tumor migration (130), invasion (130), metastasis (131), and angiogenesis (132,133), as well as in glycolysis regulation (134), the EMT (135), lipid metabolism (136) and other aspects, involving various signaling pathways (Table 1). HIF-1α promotes the migration and invasion of liver cancer cells through the IL-8/NF-κB axis. Additionally, the HIF-1α/RIT1 axis and HIF-1α/IL-8/Akt axis play important roles in facilitating the migration and invasion of human hepatoma cells. HIF-1α promotes the formation of angiogenic mimicry in HCC by upregulating expression of LOXL2 in the hypoxic tumor microenvironment. In addition, BCLAF1 promotes HCC angiogenesis by regulating HIF-1α transcription (133). HIF-1α promotes glycolysis in cancer cells through the HIF1α/PPAR-γ/PKM2 axis, leading to accelerated tumor growth, and HIF-1α-induced EMT is a critical process associated with metastasis. Feng and colleagues found that basil polysaccharide is able to inhibit hypoxia-induced metastasis and progression of liver cancer by inhibiting HIF-1α-mediated EMT. In general, reprogramming of lipid metabolism has become a hallmark of cancer, and recent studies have reported that HIF-1α is related to this process. Fatty acid-induced upregulation of FABP5 expression drives the progression of HCC through HIF-1-mediated reprogramming of lipid metabolism.
Table 1
| Function | Related genes and pathways | References |
|---|---|---|
| Migration | IL-8/NF-κB axis | (125) |
| Invasion | HIF-1α/RIT1 axis | (125) |
| Metastasis | HIF-1α/IL-8/Akt axis | (126) |
| Angiogenesis | Upregulates the expression of LOXL2 and BCLAF1 | (127,128) |
| Glycolysis | HIF-1α/PPAR-γ/PKM2 axis | (129) |
| EMT | PTEN/p-AKT/HIF-1α signaling pathway | (130) |
| Lipid metabolism | FABP5/HIF-1α axis | (131) |
| Drug resistance | miR-183-IDH2/SOCS6-HIF-1α feedback loop | (132) |
| PFKFB3/HIF-1α feedback loop | (133) |
HIF-1α, hypoxia-inducible factor-1 alpha; HCC, hepatocellular carcinoma; EMT, epithelial to mesenchymal transition.
HIF-1α is also a potential target for solving drug resistance in liver cancer. Studies have found that miR-183 regulates multidrug resistance in liver cancer cells through a miR-183-IDH2/SOCS6-HIF-1α feedback loop (137); the PFKFB3/HIF-1α feedback loop regulates sorafenib resistance in HCC cells (138). In fact, there are many studies related to hypoxia and HCC chemotherapy resistance (139-142), and increased levels of HIF-1α and vascular endothelial growth factor (VEGF) have been detected in hypoxia (26,143). Shi et al. (144) found that VEGF inhibitors can reverse the resistance of SMMC-7721 hepatoma cells to etoposide under hypoxic conditions. Mechanistically, hypoxia impedes the function of etoposide in inducing DNA damage and cell death and leads to drug resistance. Intervention of VEGF suppresses hypoxia induction by HIF-1α, reversing drug resistance in SMMC-7721 cells. In addition, Zhang and colleagues (145) found that geniposide suppresses secretion of VEGF, migration of endothelial cells, and formation of blood vessels in tumors independent of HIF-1, providing a new anti-VEGF mechanism for the treatment of HCC. In summary, overexpression of HIF-1α promotes HCC; therefore, HIF-1α inhibitors may be used to treat liver cancer. In addition, it has to be said that HIF-2α also plays an important role in HCC. One study showed that HIF-2α was associated with angiogenesis and poor prognosis in HCC (146). However, Sun et al. (147) found HIF-2α regulated autophagy and apoptosis, and high expression of HIF-2α was related to a better prognosis in HCC. Clearly, the mechanisms regulating HIF-2α function need to be further explored.
All in all, Table 2 summarizes the relationship between HIF-1α and chronic liver diseases such as ALD, NAFLD, AILI, viral hepatitis, and HCC, and the specific mechanism of action.
Table 2
| Diseases | Inhibit or promote disease progression | Key mechanism | References |
|---|---|---|---|
| ALD | Inhibit | Protect intestinal integrity | (77-79) |
| Reduce fatty acid synthesis | (80) | ||
| Promote | Cause lipid accumulation, and increase portal pressure | (81,84,86-89) | |
| Through the Akt/HIF-1α pathway | (83) | ||
| NAFLD | Inhibit | Inhibit excessive accumulation of liver fat | (90,94,95) |
| Promote | Promote liver fibrosis | (96-98) | |
| AILI | Inhibit | Contribute to hemostasis | (105) |
| Reduce inflammatory reaction | (105,106) | ||
| Improve centrilobular necrosis | (107) | ||
| Viral hepatitis | – | Increase autotaxin protein expression | (111) |
| Facilitate HIF-1α-mediated glycolytic adaptation | (112) | ||
| HCC | Promote | Promote migration, invasion, metastasis and angiogenesis | (125-127) |
HIF-1α, hypoxia-inducible factor-1 alpha; ALD, alcoholic liver disease; NAFLD, nonalcoholic fatty liver disease; AILI, acetaminophen (APAP)-induced liver injury; HCC, hepatocellular carcinoma.
Potential therapeutic value
As mentioned above, anti-PHD2 treatment significantly improves APAP-induced central lobular necrosis in AILI by upregulating HIF-1α levels (110). In addition, the systemic inactivation of 4-hydroxylase 2, the pro-product of HIF, can prevent alcohol-induced fatty liver disease (148,149). In NAFLD, HIF-P4H-2 inhibited and enhanced intestinal fructose metabolism, and induced heat generation to prevent the occurrence of NAFLD (150). PHD inhibitors act as stabilizers of HIFs in vivo (151,152). JTZ-951 inhibited PHD, which could reduce liver-related diseases in mice on a high-fat diet (151). In HCC, VEGF intervention inhibits hypoxia-induced HIF-1α, preventing drug resistance (144). Certain HIF-targeted drugs are continuously passing clinical trials. EZN-2968, an antisense oligonucleotide inhibitor of HIF-1α, is mainly used in the treatment of HCC, and clinical trials have been completed (NCT01120288) (32). In addition, the PHD inhibitor ethyl 3,4-dihydroxybenzoate has been shown to activate HIF-1α and its target HMOX1, thereby inhibiting the mitochondrial permeability transition and reducing IR-induced liver damage (153). As a potential therapeutic target, HIF-1α provides a new perspective for the treatment of various liver diseases (154,155).
Conclusions
Overall, HIF-1α is widely involved in the occurrence, development and prognosis of various liver diseases, and there is increasing evidence that HIF-1α may be involved in complex signaling pathways to regulate its own expression in a variety of liver disease processes. The results of such studies have important implications for targeting HIF-1α in treatment for liver disease. Further in-depth research on HIF-1α and liver disease is warranted.
Acknowledgments
Funding: This study was supported by grants awarded by the National Science and Technology Major Project of China (No. 2018ZX10302206), Science and Technology Major Projects of Zhejiang Province (No. 2018C04016) and Major Science and Technology Projects for Liver Disease Research Fund of Ningbo (No. 2016C51008).
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-4222/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-4222/coif). All authors report that this study was supported by grants awarded by the National Science and Technology Major Project of China (No. 2018ZX10302206), Science and Technology Major Projects of Zhejiang Province (No. 2018C04016) and Major Science and Technology Projects for Liver Disease Research Fund of Ningbo (No. 2016C51008). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Di Ciaula A, Calamita G, Shanmugam H, et al. Mitochondria Matter: Systemic Aspects of Nonalcoholic Fatty Liver Disease (NAFLD) and Diagnostic Assessment of Liver Function by Stable Isotope Dynamic Breath Tests. Int J Mol Sci 2021;22:7702. [Crossref] [PubMed]
- Kroll T, Prescher M, Smits SHJ, et al. Structure and Function of Hepatobiliary ATP Binding Cassette Transporters. Chem Rev 2021;121:5240-88. [Crossref] [PubMed]
- Wulkersdorfer B, Wanek T, Bauer M, et al. Using positron emission tomography to study transporter-mediated drug-drug interactions in tissues. Clin Pharmacol Ther 2014;96:206-13. [Crossref] [PubMed]
- Wang X, Tang H, Teng M, et al. Mapping of hepatic expression quantitative trait loci (eQTLs) in a Han Chinese population. J Med Genet 2014;51:319-26. [Crossref] [PubMed]
- Matyas C, Haskó G, Liaudet L, et al. Interplay of cardiovascular mediators, oxidative stress and inflammation in liver disease and its complications. Nat Rev Cardiol 2021;18:117-35. [Crossref] [PubMed]
- Schön MR, Kollmar O, Wolf S, et al. Liver transplantation after organ preservation with normothermic extracorporeal perfusion. Ann Surg 2001;233:114-23. [Crossref] [PubMed]
- Dietrich CG, Geier A, Wasmuth HE, et al. Influence of biliary cirrhosis on the detoxification and elimination of a food derived carcinogen. Gut 2004;53:1850-5. [Crossref] [PubMed]
- Reshetnyak VI. Primary biliary cirrhosis: Clinical and laboratory criteria for its diagnosis. World J Gastroenterol 2015;21:7683-708. [Crossref] [PubMed]
- Wu SJ, Yang YH, Tsuneyama K, et al. Innate immunity and primary biliary cirrhosis: activated invariant natural killer T cells exacerbate murine autoimmune cholangitis and fibrosis. Hepatology 2011;53:915-25. [Crossref] [PubMed]
- Ma L, Wu Y, Li Y, et al. Current Advances on 3D-Bioprinted Liver Tissue Models. Adv Healthc Mater 2020;9:e2001517. [Crossref] [PubMed]
- Limketkai BN, Mehta SH, Sutcliffe CG, et al. Relationship of liver disease stage and antiviral therapy with liver-related events and death in adults coinfected with HIV/HCV. JAMA 2012;308:370-8. [Crossref] [PubMed]
- Nassir F, Ibdah JA. Role of mitochondria in alcoholic liver disease. World J Gastroenterol 2014;20:2136-42. [Crossref] [PubMed]
- Dwyer BJ, Macmillan MT, Brennan PN, et al. Cell therapy for advanced liver diseases: Repair or rebuild. J Hepatol 2021;74:185-99. [Crossref] [PubMed]
- Acharya C, Bajaj JS. Chronic Liver Diseases and the Microbiome-Translating Our Knowledge of Gut Microbiota to Management of Chronic Liver Disease. Gastroenterology 2021;160:556-72. [Crossref] [PubMed]
- Tanwar S, Rhodes F, Srivastava A, et al. Inflammation and fibrosis in chronic liver diseases including non-alcoholic fatty liver disease and hepatitis C. World J Gastroenterol 2020;26:109-33. [Crossref] [PubMed]
- Semenza GL, Nejfelt MK, Chi SM, et al. Hypoxia-inducible nuclear factors bind to an enhancer element located 3' to the human erythropoietin gene. Proc Natl Acad Sci U S A 1991;88:5680-4. [Crossref] [PubMed]
- McGettrick AF, O'Neill LAJ. The Role of HIF in Immunity and Inflammation. Cell Metab 2020;32:524-36. [Crossref] [PubMed]
- Ladroue C, Hoogewijs D, Gad S, et al. Distinct deregulation of the hypoxia inducible factor by PHD2 mutants identified in germline DNA of patients with polycythemia. Haematologica 2012;97:9-14. [Crossref] [PubMed]
- Semenza GL. Oxygen sensing, homeostasis, and disease. N Engl J Med 2011;365:537-47. [Crossref] [PubMed]
- Laughner E, Taghavi P, Chiles K, et al. HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1alpha (HIF-1alpha) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol Cell Biol 2001;21:3995-4004. [Crossref] [PubMed]
- Semenza GL. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology (Bethesda) 2004;19:176-82. [Crossref] [PubMed]
- Sharp FR, Bernaudin M. HIF1 and oxygen sensing in the brain. Nat Rev Neurosci 2004;5:437-48. [Crossref] [PubMed]
- Nath B, Szabo G. Hypoxia and hypoxia inducible factors: diverse roles in liver diseases. Hepatology 2012;55:622-33. [Crossref] [PubMed]
- Roth KJ, Copple BL. Role of Hypoxia-Inducible Factors in the Development of Liver Fibrosis. Cell Mol Gastroenterol Hepatol 2015;1:589-97. [Crossref] [PubMed]
- Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001;292:468-72. [Crossref] [PubMed]
- Palazon A, Goldrath AW, Nizet V, et al. HIF transcription factors, inflammation, and immunity. Immunity 2014;41:518-28. [Crossref] [PubMed]
- Kietzmann T, Cornesse Y, Brechtel K, et al. Perivenous expression of the mRNA of the three hypoxia-inducible factor alpha-subunits, HIF1alpha, HIF2alpha and HIF3alpha, in rat liver. Biochem J 2001;354:531-7. [Crossref] [PubMed]
- Li HY, Yuan Y, Fu YH, et al. Hypoxia-inducible factor-1α: A promising therapeutic target for vasculopathy in diabetic retinopathy. Pharmacol Res 2020;159:104924. [Crossref] [PubMed]
- Semenza GL, Agani F, Booth G, et al. Structural and functional analysis of hypoxia-inducible factor 1. Kidney Int 1997;51:553-5. [Crossref] [PubMed]
- Luo G, Gu YZ, Jain S, et al. Molecular characterization of the murine Hif-1 alpha locus. Gene Expr 1997;6:287-99. [PubMed]
- Balamurugan K, Wang JM, Tsai HH, et al. The tumour suppressor C/EBPδ inhibits FBXW7 expression and promotes mammary tumour metastasis. EMBO J 2010;29:4106-17. [Crossref] [PubMed]
- Ju C, Colgan SP, Eltzschig HK. Hypoxia-inducible factors as molecular targets for liver diseases. J Mol Med (Berl) 2016;94:613-27. [Crossref] [PubMed]
- Lee JW, Bae SH, Jeong JW, et al. Hypoxia-inducible factor (HIF-1)alpha: its protein stability and biological functions. Exp Mol Med 2004;36:1-12. [Crossref] [PubMed]
- Bharadwaj LA, Davies GF, Xavier IJ, et al. l-carnosine and verapamil inhibit hypoxia-induced expression of hypoxia inducible factor (HIF-1 alpha) in H9c2 cardiomyoblasts. Pharmacol Res 2002;45:175-81. [Crossref] [PubMed]
- Appelhoff RJ, Tian YM, Raval RR, et al. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem 2004;279:38458-65. [Crossref] [PubMed]
- Berra E, Benizri E, Ginouvès A, et al. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J 2003;22:4082-90. [Crossref] [PubMed]
- Chua YL, Dufour E, Dassa EP, et al. Stabilization of hypoxia-inducible factor-1alpha protein in hypoxia occurs independently of mitochondrial reactive oxygen species production. J Biol Chem 2010;285:31277-84. [Crossref] [PubMed]
- Pereira T, Zheng X, Poellinger L. Degradation of the hypoxia-inducible factor 1alpha: where does it happen? Cell Cycle 2006;5:2720-2. [Crossref] [PubMed]
- Peng X, Gao H, Xu R, et al. The interplay between HIF-1α and noncoding RNAs in cancer. J Exp Clin Cancer Res 2020;39:27. [Crossref] [PubMed]
- Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer 2002;2:38-47. [Crossref] [PubMed]
- Lando D, Peet DJ, Gorman JJ, et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev 2002;16:1466-71. [Crossref] [PubMed]
- Peet D, Linke S. Regulation of HIF: asparaginyl hydroxylation. Novartis Found Symp 2006;272:37-49; discussion 49-53, 131-40.
- Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev 2001;15:2675-86. [Crossref] [PubMed]
- Jin X, Dai L, Ma Y, et al. Implications of HIF-1α in the tumorigenesis and progression of pancreatic cancer. Cancer Cell Int 2020;20:273. [Crossref] [PubMed]
- Seo HW, Kim EJ, Na H, et al. Transcriptional activation of hypoxia-inducible factor-1alpha by HDAC4 and HDAC5 involves differential recruitment of p300 and FIH-1. FEBS Lett 2009;583:55-60. [Crossref] [PubMed]
- Kang J, Chun YS, Huh J, et al. FIH permits NAA10 to catalyze the oxygen-dependent lysyl-acetylation of HIF-1α. Redox Biol 2018;19:364-74. [Crossref] [PubMed]
- Geng H, Liu Q, Xue C, et al. HIF1α protein stability is increased by acetylation at lysine 709. J Biol Chem 2012;287:35496-505. [Crossref] [PubMed]
- Seo KS, Park JH, Heo JY, et al. SIRT2 regulates tumour hypoxia response by promoting HIF-1α hydroxylation. Oncogene 2015;34:1354-62. [Crossref] [PubMed]
- Kierans SJ, Taylor CT. Regulation of glycolysis by the hypoxia-inducible factor (HIF): implications for cellular physiology. J Physiol 2021;599:23-37. [Crossref] [PubMed]
- Bracken CP, Whitelaw ML, Peet DJ. The hypoxia-inducible factors: key transcriptional regulators of hypoxic responses. Cell Mol Life Sci 2003;60:1376-93. [Crossref] [PubMed]
- Iyer NV, Kotch LE, Agani F, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev 1998;12:149-62. [Crossref] [PubMed]
- Masoud GN, Li W. HIF-1α pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B 2015;5:378-89. [Crossref] [PubMed]
- Yeo EJ. Hypoxia and aging. Exp Mol Med 2019;51:1-15. [PubMed]
- Semenza GL. Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol Med 2001;7:345-50. [Crossref] [PubMed]
- Takahashi Y, Takahashi S, Shiga Y, et al. Hypoxic induction of prolyl 4-hydroxylase alpha (I) in cultured cells. J Biol Chem 2000;275:14139-46. [Crossref] [PubMed]
- Peng T, Du SY, Son M, et al. HIF-1α is a negative regulator of interferon regulatory factors: Implications for interferon production by hypoxic monocytes. Proc Natl Acad Sci U S A 2021;118:e2106017118. [Crossref] [PubMed]
- Shin DH, Lin H, Zheng H, et al. HIF-1α-mediated upregulation of TASK-2 K+ channels augments Ca2+ signaling in mouse B cells under hypoxia. J Immunol 2014;193:4924-33. [Crossref] [PubMed]
- Melstrom LG, Salabat MR, Ding XZ, et al. Apigenin down-regulates the hypoxia response genes: HIF-1α, GLUT-1, and VEGF in human pancreatic cancer cells. J Surg Res 2011;167:173-81. [Crossref] [PubMed]
- Voit RA, Sankaran VG. Stabilizing HIF to Ameliorate Anemia. Cell 2020;180:6. [Crossref] [PubMed]
- Nadai T, Narumi K, Furugen A, et al. Pharmacological Inhibition of MCT4 Reduces 4-Hydroxytamoxifen Sensitivity by Increasing HIF-1α Protein Expression in ER-Positive MCF-7 Breast Cancer Cells. Biol Pharm Bull 2021;44:1247-53. [Crossref] [PubMed]
- Martinez-Outschoorn U, Sotgia F, Lisanti MP. Tumor microenvironment and metabolic synergy in breast cancers: critical importance of mitochondrial fuels and function. Semin Oncol 2014;41:195-216. [Crossref] [PubMed]
- Luo F, Zou Z, Liu X, et al. Enhanced glycolysis, regulated by HIF-1α via MCT-4, promotes inflammation in arsenite-induced carcinogenesis. Carcinogenesis 2017;38:615-26. [Crossref] [PubMed]
- Lai SW, Lin HJ, Liu YS, et al. Monocarboxylate Transporter 4 Regulates Glioblastoma Motility and Monocyte Binding Ability. Cancers (Basel) 2020;12:380. [Crossref] [PubMed]
- Silagi ES, Schoepflin ZR, Seifert EL, et al. Bicarbonate Recycling by HIF-1-Dependent Carbonic Anhydrase Isoforms 9 and 12 Is Critical in Maintaining Intracellular pH and Viability of Nucleus Pulposus Cells. J Bone Miner Res 2018;33:338-55. [Crossref] [PubMed]
- Ni HM, Bhakta A, Wang S, et al. Role of hypoxia inducing factor-1β in alcohol-induced autophagy, steatosis and liver injury in mice. PLoS One 2014;9:e115849. [Crossref] [PubMed]
- Parker R, Kim SJ, Gao B. Alcohol, adipose tissue and liver disease: mechanistic links and clinical considerations. Nat Rev Gastroenterol Hepatol 2018;15:50-9. [Crossref] [PubMed]
- Zhang P, Qiang X, Zhang M, et al. Demethyleneberberine, a natural mitochondria-targeted antioxidant, inhibits mitochondrial dysfunction, oxidative stress, and steatosis in alcoholic liver disease mouse model. J Pharmacol Exp Ther 2015;352:139-47. [Crossref] [PubMed]
- Seitz HK, Bataller R, Cortez-Pinto H, et al. Publisher Correction: Alcoholic liver disease. Nat Rev Dis Primers 2018;4:18. [Crossref] [PubMed]
- Bajaj JS. Alcohol, liver disease and the gut microbiota. Nat Rev Gastroenterol Hepatol 2019;16:235-46. [Crossref] [PubMed]
- Liangpunsakul S, Haber P, McCaughan GW. Alcoholic Liver Disease in Asia, Europe, and North America. Gastroenterology 2016;150:1786-97. [Crossref] [PubMed]
- Fernandes LMP, Lopes KS, Santana LNS, et al. Repeated Cycles of Binge-Like Ethanol Intake in Adolescent Female Rats Induce Motor Function Impairment and Oxidative Damage in Motor Cortex and Liver, but Not in Blood. Oxid Med Cell Longev 2018;2018:3467531. [Crossref] [PubMed]
- Ceni E, Mello T, Galli A. Pathogenesis of alcoholic liver disease: role of oxidative metabolism. World J Gastroenterol 2014;20:17756-72. [Crossref] [PubMed]
- Rath E, Moschetta A, Haller D. Mitochondrial function - gatekeeper of intestinal epithelial cell homeostasis. Nat Rev Gastroenterol Hepatol 2018;15:497-516. [Crossref] [PubMed]
- Louvet A, Mathurin P. Alcoholic liver disease: mechanisms of injury and targeted treatment. Nat Rev Gastroenterol Hepatol 2015;12:231-42. [Crossref] [PubMed]
- Arteel GE, Iimuro Y, Yin M, et al. Chronic enteral ethanol treatment causes hypoxia in rat liver tissue in vivo. Hepatology 1997;25:920-6. [Crossref] [PubMed]
- Zelickson BR, Benavides GA, Johnson MS, et al. Nitric oxide and hypoxia exacerbate alcohol-induced mitochondrial dysfunction in hepatocytes. Biochim Biophys Acta 2011;1807:1573-82. [Crossref] [PubMed]
- Lin H, Chen D, Du Q, et al. Dietary Copper Plays an Important Role in Maintaining Intestinal Barrier Integrity During Alcohol-Induced Liver Disease Through Regulation of the Intestinal HIF-1α Signaling Pathway and Oxidative Stress. Front Physiol 2020;11:369. [Crossref] [PubMed]
- Shao T, Zhao C, Li F, et al. Intestinal HIF-1α deletion exacerbates alcoholic liver disease by inducing intestinal dysbiosis and barrier dysfunction. J Hepatol 2018;69:886-95. [Crossref] [PubMed]
- Wang Y, Kirpich I, Liu Y, et al. Lactobacillus rhamnosus GG treatment potentiates intestinal hypoxia-inducible factor, promotes intestinal integrity and ameliorates alcohol-induced liver injury. Am J Pathol 2011;179:2866-75. [Crossref] [PubMed]
- Nishiyama Y, Goda N, Kanai M, et al. HIF-1α induction suppresses excessive lipid accumulation in alcoholic fatty liver in mice. J Hepatol 2012;56:441-7. [Crossref] [PubMed]
- Li L, Chen SH, Zhang Y, et al. Is the hypoxia-inducible factor-1 alpha mRNA expression activated by ethanol-induced injury, the mechanism underlying alcoholic liver disease? Hepatobiliary Pancreat Dis Int 2006;5:560-3. [PubMed]
- Zhou JY, Jiang ZA, Zhao CY, et al. Long-term binge and escalating ethanol exposure causes necroinflammation and fibrosis in rat liver. Alcohol Clin Exp Res 2013;37:213-22. [Crossref] [PubMed]
- Wang Z, Li B, Jiang H, et al. IL-8 exacerbates alcohol-induced fatty liver disease via the Akt/HIF-1α pathway in human IL-8-expressing mice. Cytokine 2021;138:155402. [Crossref] [PubMed]
- Satishchandran A, Ambade A, Rao S, et al. MicroRNA 122, Regulated by GRLH2, Protects Livers of Mice and Patients From Ethanol-Induced Liver Disease. Gastroenterology 2018;154:238-252.e7. [Crossref] [PubMed]
- Wang X, Wu D, Yang L, et al. Cytochrome P450 2E1 potentiates ethanol induction of hypoxia and HIF-1α in vivo. Free Radic Biol Med 2013;63:175-86. [Crossref] [PubMed]
- Mehal WZ. HIF-1α is a major and complex player in alcohol induced liver diseases. J Hepatol 2012;56:311-2. [Crossref] [PubMed]
- Nath B, Levin I, Csak T, et al. Hepatocyte-specific hypoxia-inducible factor-1α is a determinant of lipid accumulation and liver injury in alcohol-induced steatosis in mice. Hepatology 2011;53:1526-37. [Crossref] [PubMed]
- Jin H, Lian N, Bian M, et al. Oroxylin A prevents alcohol-induced hepatic steatosis through inhibition of hypoxia inducible factor 1alpha. Chem Biol Interact 2018;285:14-20. [Crossref] [PubMed]
- Guo X, Wang Y, Shen Y, et al. Gene expression profiles of sodium-dependent vitamin C transporters in mice after alcohol consumption. Acta Biochim Biophys Sin (Shanghai) 2013;45:912-20. [Crossref] [PubMed]
- Arai T, Tanaka M, Goda N. HIF-1-dependent lipin1 induction prevents excessive lipid accumulation in choline-deficient diet-induced fatty liver. Sci Rep 2018;8:14230. [Crossref] [PubMed]
- Targher G, Chonchol M, Zoppini G, et al. Risk of chronic kidney disease in patients with non-alcoholic fatty liver disease: is there a link? J Hepatol 2011;54:1020-9. [Crossref] [PubMed]
- Henao-Mejia J, Elinav E, Jin C, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 2012;482:179-85. [Crossref] [PubMed]
- Finck BN, Gropler MC, Chen Z, et al. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab 2006;4:199-210. [Crossref] [PubMed]
- He Y, Yang W, Gan L, et al. Silencing HIF-1α aggravates non-alcoholic fatty liver disease in vitro through inhibiting PPAR-α/ANGPTL4 singling pathway. Gastroenterol Hepatol 2021;44:355-65. [Crossref] [PubMed]
- Yoo W, Noh KH, Ahn JH, et al. HIF-1α expression as a protective strategy of HepG2 cells against fatty acid-induced toxicity. J Cell Biochem 2014;115:1147-58. [Crossref] [PubMed]
- Mesarwi OA, Shin MK, Bevans-Fonti S, et al. Hepatocyte Hypoxia Inducible Factor-1 Mediates the Development of Liver Fibrosis in a Mouse Model of Nonalcoholic Fatty Liver Disease. PLoS One 2016;11:e0168572. [Crossref] [PubMed]
- Han J, He Y, Zhao H, et al. Hypoxia inducible factor-1 promotes liver fibrosis in nonalcoholic fatty liver disease by activating PTEN/p65 signaling pathway. J Cell Biochem 2019;120:14735-44. [Crossref] [PubMed]
- Liu X, Huang K, Niu Z, et al. Protective effect of isochlorogenic acid B on liver fibrosis in non-alcoholic steatohepatitis of mice. Basic Clin Pharmacol Toxicol 2019;124:144-53. [Crossref] [PubMed]
- Rankin EB, Rha J, Selak MA, et al. Hypoxia-inducible factor 2 regulates hepatic lipid metabolism. Mol Cell Biol 2009;29:4527-38. [Crossref] [PubMed]
- Morello E, Sutti S, Foglia B, et al. Hypoxia-inducible factor 2α drives nonalcoholic fatty liver progression by triggering hepatocyte release of histidine-rich glycoprotein. Hepatology 2018;67:2196-214. [Crossref] [PubMed]
- Xie C, Yagai T, Luo Y, et al. Activation of intestinal hypoxia-inducible factor 2α during obesity contributes to hepatic steatosis. Nat Med 2017;23:1298-308. [Crossref] [PubMed]
- Platko K, Lebeau PF, Byun JH, et al. GDF10 blocks hepatic PPARγ activation to protect against diet-induced liver injury. Mol Metab 2019;27:62-74. [Crossref] [PubMed]
- Walesky CM, Kolb KE, Winston CL, et al. Functional compensation precedes recovery of tissue mass following acute liver injury. Nat Commun 2020;11:5785. [Crossref] [PubMed]
- Kusumanchi P, Liang T, Zhang T, et al. Stress-Responsive Gene FK506-Binding Protein 51 Mediates Alcohol-Induced Liver Injury Through the Hippo Pathway and Chemokine (C-X-C Motif) Ligand 1 Signaling. Hepatology 2021;74:1234-50. [Crossref] [PubMed]
- Lucena MI, Sanabria J, García-Cortes M, et al. Drug-induced liver injury in older people. Lancet Gastroenterol Hepatol 2020;5:862-74. [Crossref] [PubMed]
- Andrade RJ, Chalasani N, Björnsson ES, et al. Drug-induced liver injury. Nat Rev Dis Primers 2019;5:58. [Crossref] [PubMed]
- Song B, Zhang C, Hu W, et al. Nano-designed carbon monoxide donor SMA/CORM2 exhibits protective effect against acetaminophen induced liver injury through macrophage reprograming and promoting liver regeneration. J Control Release 2021;331:350-63. [Crossref] [PubMed]
- Sparkenbaugh EM, Saini Y, Greenwood KK, et al. The role of hypoxia-inducible factor-1α in acetaminophen hepatotoxicity. J Pharmacol Exp Ther 2011;338:492-502. [Crossref] [PubMed]
- Suzuki T, Minagawa S, Yamazaki T, et al. Loss of hypoxia inducible factor-1α aggravates γδ T-cell-mediated inflammation during acetaminophen-induced liver injury. Hepatol Commun 2018;2:571-81. [Crossref] [PubMed]
- Zhao L, Zhao J, Gao R, et al. Intrabody against prolyl hydroxylase 2 ameliorates acetaminophen-induced acute liver injury in mice via concomitant promotion of angiogenesis and redox homeostasis. Biomed Pharmacother 2020;123:109783. [Crossref] [PubMed]
- Tse AP, Sze KM, Shea QT, et al. Hepatitis transactivator protein X promotes extracellular matrix modification through HIF/LOX pathway in liver cancer. Oncogenesis 2018;7:44. [Crossref] [PubMed]
- Liu LP, Hu BG, Ye C, et al. HBx mutants differentially affect the activation of hypoxia-inducible factor-1α in hepatocellular carcinoma. Br J Cancer 2014;110:1066-73. [Crossref] [PubMed]
- Moon EJ, Jeong CH, Jeong JW, et al. Hepatitis B virus X protein induces angiogenesis by stabilizing hypoxia-inducible factor-1alpha. FASEB J 2004;18:382-4. [Crossref] [PubMed]
- Farquhar MJ, Humphreys IS, Rudge SA, et al. Autotaxin-lysophosphatidic acid receptor signalling regulates hepatitis C virus replication. J Hepatol 2017;66:919-29. [Crossref] [PubMed]
- Ripoli M, D'Aprile A, Quarato G, et al. Hepatitis C virus-linked mitochondrial dysfunction promotes hypoxia-inducible factor 1 alpha-mediated glycolytic adaptation. J Virol 2010;84:647-60. [Crossref] [PubMed]
- Wilson GK, Brimacombe CL, Rowe IA, et al. A dual role for hypoxia inducible factor-1α in the hepatitis C virus lifecycle and hepatoma migration. J Hepatol 2012;56:803-9. [Crossref] [PubMed]
- Jiang Y, Sun A, Zhao Y, et al. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature 2019;567:257-61. [Crossref] [PubMed]
- Shibata T, Aburatani H. Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol 2014;11:340-9. [Crossref] [PubMed]
- Ho DWH, Chan LK, Chiu YT, et al. TSC1/2 mutations define a molecular subset of HCC with aggressive behaviour and treatment implication. Gut 2017;66:1496-506. [Crossref] [PubMed]
- Reichl P, Dengler M, van Zijl F, et al. Axl activates autocrine transforming growth factor-β signaling in hepatocellular carcinoma. Hepatology 2015;61:930-41. [Crossref] [PubMed]
- Meng X, Franklin DA, Dong J, et al. MDM2-p53 pathway in hepatocellular carcinoma. Cancer Res 2014;74:7161-7. [Crossref] [PubMed]
- Gong J, Shen S, Yang Y, et al. Inhibition of FASN suppresses migration, invasion and growth in hepatoma carcinoma cells by deregulating the HIF-1α/IGFBP1 pathway. Int J Oncol 2017;50:883-92. [Crossref] [PubMed]
- Abdel-Rahman O, Elsayed Z. External beam radiotherapy for unresectable hepatocellular carcinoma. Cochrane Database Syst Rev 2017;3:CD011314. [Crossref] [PubMed]
- Ramanathan R, Sharma A, Lee DD, et al. Multimodality therapy and liver transplantation for hepatocellular carcinoma: a 14-year prospective analysis of outcomes. Transplantation 2014;98:100-6. [Crossref] [PubMed]
- Shao G, Zou Y, Lucatelli P, et al. Chinese expert consensus on technical recommendations for the standard operation of drug-eluting beads for transvascular embolization. Ann Transl Med 2021;9:714. [Crossref] [PubMed]
- Shah JL, Zendejas-Ruiz IR, Thornton LM, et al. Neoadjuvant transarterial radiation lobectomy for colorectal hepatic metastases: a small cohort analysis on safety, efficacy, and radiopathologic correlation. J Gastrointest Oncol 2017;8:E43-51. [Crossref] [PubMed]
- Niu L, Liu L, Yang S, et al. New insights into sorafenib resistance in hepatocellular carcinoma: Responsible mechanisms and promising strategies. Biochim Biophys Acta Rev Cancer 2017;1868:564-70. [Crossref] [PubMed]
- Zheng SS, Chen XH, Yin X, et al. Prognostic significance of HIF-1α expression in hepatocellular carcinoma: a meta-analysis. PLoS One 2013;8:e65753. [Crossref] [PubMed]
- Yang SL, Liu LP, Jiang JX, et al. The correlation of expression levels of HIF-1α and HIF-2α in hepatocellular carcinoma with capsular invasion, portal vein tumor thrombi and patients' clinical outcome. Jpn J Clin Oncol 2014;44:159-67. [Crossref] [PubMed]
- Feng W, Xue T, Huang S, et al. HIF-1α promotes the migration and invasion of hepatocellular carcinoma cells via the IL-8-NF-κB axis. Cell Mol Biol Lett 2018;23:26. [Crossref] [PubMed]
- Feng B, Zhu Y, Su Z, et al. Basil polysaccharide attenuates hepatocellular carcinoma metastasis in rat by suppressing H3K9me2 histone methylation under hepatic artery ligation-induced hypoxia. Int J Biol Macromol 2018;107:2171-9. [Crossref] [PubMed]
- Wang M, Zhao X, Zhu D, et al. HIF-1α promoted vasculogenic mimicry formation in hepatocellular carcinoma through LOXL2 up-regulation in hypoxic tumor microenvironment. J Exp Clin Cancer Res 2017;36:60. [Crossref] [PubMed]
- Wen Y, Zhou X, Lu M, et al. Bclaf1 promotes angiogenesis by regulating HIF-1α transcription in hepatocellular carcinoma. Oncogene 2019;38:1845-59. [Crossref] [PubMed]
- Feng J, Dai W, Mao Y, et al. Simvastatin re-sensitizes hepatocellular carcinoma cells to sorafenib by inhibiting HIF-1α/PPAR-γ/PKM2-mediated glycolysis. J Exp Clin Cancer Res 2020;39:24. [Crossref] [PubMed]
- Feng B, Zhu Y, Sun C, et al. Basil polysaccharide inhibits hypoxia-induced hepatocellular carcinoma metastasis and progression through suppression of HIF-1α-mediated epithelial-mesenchymal transition. Int J Biol Macromol 2019;137:32-44. [Crossref] [PubMed]
- Seo J, Jeong DW, Park JW, et al. Fatty-acid-induced FABP5/HIF-1 reprograms lipid metabolism and enhances the proliferation of liver cancer cells. Commun Biol 2020;3:638. [Crossref] [PubMed]
- Wang XJ, Zhang DL, Fu C, et al. MiR-183 modulates multi-drug resistance in hepatocellular cancer (HCC) cells via miR-183-IDH2/SOCS6-HIF-1α feedback loop. Eur Rev Med Pharmacol Sci 2016;20:2020-7. [PubMed]
- Long Q, Zou X, Song Y, et al. PFKFB3/HIF-1α feedback loop modulates sorafenib resistance in hepatocellular carcinoma cells. Biochem Biophys Res Commun 2019;513:642-50. [Crossref] [PubMed]
- Lin Z, Niu Y, Wan A, et al. RNA m6 A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J 2020;39:e103181. [Crossref] [PubMed]
- Yu G, Chen X, Chen S, et al. Arsenic trioxide reduces chemo-resistance to 5-fluorouracil and cisplatin in HBx-HepG2 cells via complex mechanisms. Cancer Cell Int 2015;15:116. [Crossref] [PubMed]
- Fan L, Sun G, Ma T, et al. Melatonin reverses tunicamycin-induced endoplasmic reticulum stress in human hepatocellular carcinoma cells and improves cytotoxic response to doxorubicin by increasing CHOP and decreasing survivin. J Pineal Res 2013;55:184-94. [Crossref] [PubMed]
- Wu H, Wang T, Liu Y, et al. Mitophagy promotes sorafenib resistance through hypoxia-inducible ATAD3A dependent Axis. J Exp Clin Cancer Res 2020;39:274. [Crossref] [PubMed]
- Flammer J, Konieczka K, Bruno RM, et al. The eye and the heart. Eur Heart J 2013;34:1270-8. [Crossref] [PubMed]
- Shi S, Yuan C, Zhuang K, et al. Resistance of SMMC-7721 hepatoma cells to etoposide in hypoxia is reversed by VEGF inhibitor. Mol Med Rep 2015;11:3842-7. [Crossref] [PubMed]
- Zhang C, Wang N, Tan HY, et al. Direct inhibition of the TLR4/MyD88 pathway by geniposide suppresses HIF-1α-independent VEGF expression and angiogenesis in hepatocellular carcinoma. Br J Pharmacol 2020;177:3240-57. [Crossref] [PubMed]
- Bangoura G, Liu ZS, Qian Q, et al. Prognostic significance of HIF-2alpha/EPAS1 expression in hepatocellular carcinoma. World J Gastroenterol 2007;13:3176-82. [Crossref] [PubMed]
- Sun HX, Xu Y, Yang XR, et al. Hypoxia inducible factor 2 alpha inhibits hepatocellular carcinoma growth through the transcription factor dimerization partner 3/ E2F transcription factor 1-dependent apoptotic pathway. Hepatology 2013;57:1088-97. [Crossref] [PubMed]
- Laitakari A, Ollonen T, Kietzmann T, et al. Systemic inactivation of hypoxia-inducible factor prolyl 4-hydroxylase 2 in mice protects from alcohol-induced fatty liver disease. Redox Biol 2019;22:101145. [Crossref] [PubMed]
- Minamishima YA, Kaelin WG Jr. Reactivation of hepatic EPO synthesis in mice after PHD loss. Science 2010;329:407. [Crossref] [PubMed]
- Laitakari A, Tapio J, Mäkelä KA, et al. HIF-P4H-2 inhibition enhances intestinal fructose metabolism and induces thermogenesis protecting against NAFLD. J Mol Med (Berl) 2020;98:719-31. [Crossref] [PubMed]
- Saito H, Tanaka T, Sugahara M, et al. Inhibition of prolyl hydroxylase domain (PHD) by JTZ-951 reduces obesity-related diseases in the liver, white adipose tissue, and kidney in mice with a high-fat diet. Lab Invest 2019;99:1217-32. [Crossref] [PubMed]
- Taniguchi CM, Finger EC, Krieg AJ, et al. Cross-talk between hypoxia and insulin signaling through Phd3 regulates hepatic glucose and lipid metabolism and ameliorates diabetes. Nat Med 2013;19:1325-30. [Crossref] [PubMed]
- Zhong Z, Ramshesh VK, Rehman H, et al. Activation of the oxygen-sensing signal cascade prevents mitochondrial injury after mouse liver ischemia-reperfusion. Am J Physiol Gastrointest Liver Physiol 2008;295:G823-32. [Crossref] [PubMed]
- Bogaerts E, Paridaens A, Verhelst X, et al. Effect of prolyl hydroxylase domain 2 haplodeficiency on liver progenitor cell characteristics in early mouse hepatocarcinogenesis. EXCLI J 2016;15:687-98. [PubMed]
- Moon JO, Welch TP, Gonzalez FJ, et al. Reduced liver fibrosis in hypoxia-inducible factor-1alpha-deficient mice. Am J Physiol Gastrointest Liver Physiol 2009;296:G582-92. [Crossref] [PubMed]


