
A narrative review of liver regeneration—from models to molecular basis
Introduction
Liver regeneration refers to the process by which the damaged liver can repair and grow from the remaining hepatic tissue to replace the lost tissue (1). When the liver is physically damaged such as surgery or chemically damaged by alcohol or drugs, the regeneration of the liver will be stimulated and activated. Although the liver has an excellent regenerative ability, this capacity becomes overwhelmed after acute injury or resection, especially in cases of severe acute/chronic liver injury with fibrosis and abnormal hepatic structure. In the clinic, these conditions usually result in high rates of morbidity and mortality. There is a delicate balance between liver regeneration and liver damage. In other words, when the damage is within the compensatory capacity of liver regeneration, the damage stimulates the regeneration of the liver; however, when the damage exceeds the compensatory capacity of liver regeneration, it will seriously threaten the survival of the host. Therefore, in the case of decompensated liver regeneration, it is necessary to improve the understanding of liver regeneration and to innovate new clinical treatment therapies. The mechanism of liver regeneration has been the subject of research over many years and various signal pathways have been discovered. A number of treatments for liver injury regeneration have been proposed, including cellular therapy including mesenchymal stem cells (MSCs) (2), hepatic progenitor cells (HPCs) (3), and the administration of various cytokines (4). However, knowing the mechanisms facilitating the regeneration of an abnormal liver remains challenging (5). Understanding the molecular basis relevant to regeneration is a vital goal. The lessons learned from liver regeneration models are significant and help in deepening our understanding of the pathogenesis, developing novel drugs, and determining comprehensive treatments of hepatic diseases.
Liver regeneration is one of the most enigmatic and fascinating phenomena of the human organism. Although there have been similar reviews of liver regeneration before, researchers have conducted in-depth research on the factors that affect regeneration and have provided different models for regeneration with respective characteristics. This review not only comprehensively explains the advantages and disadvantages of different liver regeneration animal models and analyzes the features of each model, but also summarizes the latest results of liver regeneration mechanism research to further increase the understanding of the liver regeneration process and discusses its underlying mechanism in hepatic repair and helps us to better consider the impediments to regeneration, which may provide a more detailed insight into research and clinical therapy of liver failure.
We present the following article in accordance with the Narrative Review reporting checklist (available at https://dx.doi.org/10.21037/atm-21-5234).
Models for liver regeneration
The partial hepatectomy (PHx) model was first described in 1931 and is still a widely used animal model for liver regeneration. Briefly, the two main characteristics of this model are easy control and a regenerative environment. In addition, there are now several chemical damage models. These chemical drugs are not only accompanied by a regeneration response, but also activate an inflammatory response in the process of causing liver cell damage and death. This makes the animal model closer to the regenerative response that occurs in human liver disease. At the same time, because the reproducibility of the chemical damage model is stronger than that of PHx, it is more suitable for the study of liver regeneration in chronic liver injury. Finally, there is an emerging modeling method, the transgenic model. Compared with the other two modeling methods, its operation is simpler and more suitable for the study of specific cytokines and genes related to liver regeneration. We will summarize these reported liver regeneration models and respectively clarify their characteristics, mechanism, advantages, and methods (Figure 1).
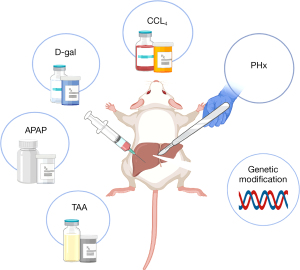
PHx
The liver regeneration induced by PHx mainly depends on the size of the functional liver resected (6). The rat model of 2/3 hepatectomy designed by Anderson and Higgins many years ago has been widely accepted (7). The advantage of 2/3 hepatectomy in rats to induce liver regeneration is that the anatomical structure of the rat is uniform, and the operator can repeat the resection in different proportions with high precision (8). Also, since the degree of damage of the model is proportional to the size of the cut liver lobe, the model is easily scalable. Regeneration is compensated solely by hypertrophy without cell division after 30% PHx, and hypertrophy precedes proliferation after 70% PHx (9). In addition, as the remaining liver cells are normal and the environment in which regeneration occurs is simple and can be used to study the time and degree of influence of different variables. The liver immediately begins to regenerate after being damaged. Within 16 hours of liver resection in rats, deoxyribonucleic acid (DNA) replication begins. In the classic model of 70% hepatectomy, the remaining part of the liver compensatorily proliferates to 45% of the original liver mass after 24 hours resection, 70% after 72 hours, 93% after 7–14 days, and basically returns to the original liver mass at approximately 20 days (1). The process of liver regeneration in mammals is similar to that in humans, and some conclusions obtained from animal models can also be applied to the study of the human liver (10). At present, PHx is the primary model for studying cytokines and signal pathways related to liver regeneration (11-13).
Carbon tetrachloride (CCl4)
The CCl4 model of liver injury in mice is the most frequently model of repeated liver damage. After CCl4-mediated damage, firstly, there is predictable parenchymal necrosis around the central vein, which peaks in 24 hours, and then liver regeneration (5). Long-term administration of CCl4 can continuously activate quiescent hepatic stellate cells (HSCs) into collagen-I producing myofibroblasts, which promotes the formation of fibrous scars (14). Failure to be degraded collagen-I severely damages HSCs apoptosis and may hinder the effective restoration of hepatocyte (15). Cessation of CCl4 administration often results in fibrosis resolution and regeneration of the liver parenchyma (16). It is known that macrophages in collagen scar regression play a critical role in liver regeneration (17), and new findings have shown that lineage-specific transcription factors are also pivotal in the progress (14).
D-galactosamine (D-gal)
D-gal inducing hepatotoxic injury has also become a common model of acute liver injury. D-gal is a disruptor of uridine triphosphate (UTP) in liver cells, which can cause diffuse hepatic necrosis and inflammation similar to viral hepatitis. Compared with other drugs, D-gal has the advantages of easier dosage control and better reproducibility (18). In the D-gal-sensitized mice, tumor necrosis factor α (TNF-α) as the main mediator participates in the entire regeneration process. It induces hepatocyte apoptosis in the early stage of acute liver injury and neutrophil migration in the later stage (19,20). D-gal is generally injected through the abdominal cavity and external jugular vein. At the same time, an animal model induced by D-gal is established by observing its clinical manifestations and survival time, detecting changes in inflammatory factor, liver function levels and histopathology (21-23). The livers of D-gal-induced mice shows spotty necrosis, lymphocyte infiltration and balloon degeneration at 6 h and 24 h, and recovery at 72 h (24).
Acetaminophen (APAP)
Since APAP is the most used analgesic in clinical practice, acute liver failure (ALF) caused by APAP intoxication is also relatively common. At present, overuse of APAP in Western countries is the main cause of ALF, accounting for almost half of all ALF cases (25). The metabolism and poisoning mechanism of APAP-induced liver failure animal model is close to clinical practice. N-acetyl-p-benzoquinone imine (NAPQI) is a reactive metabolite that binds to cellular mitochondrial proteins, causing a large number of mitochondrial oxidative dysfunction/damage and liver cell necrosis, thereby triggering APAP toxicity (26). Liver regeneration after APAP is dose- and time-dependent, and the progress is complex, involving growth factors, cytokines, angiogenic factors, and other mitogenic pathways (27). APAP is well absorbed and usually administrated by intraperitoneal injection (28-30). However, the disadvantage of this method is that due to low drug solubility, the dose concentration used in modeling is higher than the solubility at a regular temperature.
Thioacetamide (TAA)
Many studies have found that TAA can resulting in pathological changes in the liver. As a well-known hepatocarcinogen, TAA can cause different degrees of liver damage according to the time and dose of administration. Severe perivenous necrosis is the main feature of acute liver injury caused by TAA of necrotic-genic dose, followed by regeneration of hepatocytes, which provides a useful model for studying hepatocellular proliferation in respond to chemical damage (31,32). Fernández-Martínez et al. showed that hepatocytes extracted from TAA-treated mice express cyclooxygenase-2 (COX-2) protein and nitric oxide synthase-2 (NOS-2) which are involved in the initiation of regeneration after acute liver injury. Studies have found that COX-2 inhibition seems to alleviate liver injury, and loss of NOS-2 delays hepatocytes regeneration (33).
Genetically modified animals
It is challenging to replicate the features of human liver using any animal model induced by PHx or chemical materials. Therefore, genetically modified animals have been put forward as new models of liver regeneration. To some extent, these genetically modified animals are immune-deficient. In a mutant liver, fumarylacetoacetate hydrolase (Fah)-positive hepatocytes tend to have a growth advantage and widely repopulate the damaged liver. Fah-knockout mice have served as a container that can be transplanted human hepatocytes, creating “mice with human liver” (34). These chimeric animals have human-special biological functions due to human hepatic tissue and cell, making them more suitable to study human liver injury and regeneration (35).
Triggers of liver regeneration after PHx
There may be differences in the triggering causes of liver regeneration activation for different modeling methods. We will mainly explain liver regeneration triggered after PHx on account of its widespread application. The activation of cell proliferation in the process of liver regeneration first requires the cells to feel the existence of liver damage. The commonly recognized trigger factors are the hemodynamic changes of portal vein blood flow and the increase of shear stress, innate immune response, and hemostasis activation.
Elevation of shear stress
The hepatic portal vein is the main blood supply route in the liver. After 2/3 of the liver is removed, the blood in the portal vein that should flow to the whole liver only flows to the remaining 1/3 of the liver tissue (36). A simple mathematical deduction demonstrates that this will inevitably lead to two results: first, the friction exerted by blood flow on the endothelial surface increases significantly, that is, there is an increase in shear stress (37,38); second, each liver cell receiving a number of signal factors from the portal vein is several times that before liver resection.
The hepatic-portal shunt model was established to keep the blood pressure constant and stable after PHx. Previous findings indicate that the liver could not regenerate in time, which confirm the important role of portal blood pressure changes for liver injury perception and growth signal activation (39). Studies have found that hemodynamic changes in the portal vein lead to increased shear stress in liver sinusoidal endothelial cells (LSECs), which in turn promotes the release of nitric oxide (NO), which increases the sensitivity of hepatocytes to hepatocyte growth factor (HGF) (40), induces vascular endothelial growth factor (VEGF) (41,42), and stimulates HSCs to release HGF and VEGF (43). The interleukin (IL)-6 released by LSEC may also lead to an increase in shear stress. Compared with unstretched LSECs, mechanically stretched LSECs releases more IL-6 (44).
Correspondingly, an improvement in shear stress will increase the activity of urokinase-type plasminogen activator (uPA) (45,46). The rapid activation of uPA causes the conversion of plasminogen to plasmin, which subsequently initiates breakdown of extracellular matrix (ECM) constituents and cuts precursor (pro-HGF) molecules into active HGF binding to hepatocyte growth factor receptor (HGFR or c-Met) (47-50). EGF increases in relative concentration due to the increase in portal venous flow and motivates the epidermal growth factor receptor (EGFR, also known as ErbB) (51,52). Activated HGFR and EGFR trigger the liver regeneration cascade, including phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) and mitogen-activated protein kinases (MAPK, also known as Ras/Raf/MEK/Erk), and elevate the enhanced expression of c-myc, c-fos, c-jun, and other transcription factors, which finally facilitates protein synthesis and cell division (40).
Innate immune response
The innate immune response is also regarded as a major stimulus of liver regeneration (53,54). As components of innate immunity, lipopolysaccharide (LPS) and complements (such as C3a and C5a) are released from the intestinal tract into the portal vein bloodstream, and activate macrophages (or namely Kupffer cells, KCs) by binding to Toll-like receptor 4 (TLR4) and complement receptor, respectively (55-58). These interactions lead to the stimulation of an important signaling pathway, known as the nuclear factor kappa B (NF-kB) pathway (59). As a dimeric transcription factor, NF-kB is composed of seven different proteins: NF-κB1 (p105 and p50), NF-κB2 (p100 and p52), RelA (p65), c-Rel and RelB (60). Under normal circumstances, NF-kB binds to its inhibitory KB protein (IKB) and is accumulated in the cytoplasm of KCs in the form of a complex. When KCs are stimulated, IKB is phosphorylated and degraded so that NF-kB is released into the nucleus (61), thereby triggering the release of TNF-α and IL-6 (62-64).
Increase of shear stress or innate immune response is a “double-edged sword” in liver regeneration. When the hepatectomy area is hard to compensate, the shear stress will cause hepatocyte damage and death, which is also called post-hepatectomy liver failure (65). An overly strong immune response will not only not promote liver regeneration, but will also aggravate both liver damage and the condition (60,66).
Hemostasis activation
Hemostasis is not only the key to a good prognosis after PHx, but is also associated to liver regeneration (67). Many hemostatic factors have been reported to be involved in liver regeneration, among which platelets undoubtedly play an important role in this process (68,69). Studies have shown that the lower the platelet count, the worse the liver regeneration (70,71). After PHx, platelets quickly migrate to the Disse space, release their contents, and stimulate the proliferation of hepatocytes or LSEC through HGF, VEGF, or serotonin (72-74). Serotonin has been clearly able to promote liver regeneration (75,76) and the mechanism may be related to the Hippo signaling pathway (77). In addition, platelet promotion of liver regeneration may also be related to their mobilization of bone marrow mesenchymal cells to migrate to the damaged liver (78) (Table 1).
Table 1
| Trigger | Animal | Degree of PHx | Effect | Mechanism | Ref |
|---|---|---|---|---|---|
| Elevation of shear stress | Rat | 2/3PHx | Initiates and maintains liver regeneration | Proper portal blood perfusion; Hepatocyte membrane and sodium-potassium pump changes | (38) |
| Elevation of shear stress | Rat | 2/3PHx | Triggers the liver regeneration cascade | Expression of c-fos mRNA↑; Release of NO and proliferation factors↑ | (40) |
| Elevation of shear stress | Mice | 2/3PHx | The decreased serum nitrate and nitrite levels lead to lower liver mass recovery and higher ALT | Release of NO↓; The HSP70 family and Ki-67↓; Induction of Nrp1 and EGFR↓ | (42) |
| Elevation of shear stress | Rat | 2/3PHx | Initiates liver regeneration | uPA and uPAR activation; Induction of active HGF↑ | (46) |
| Innate immune response | Mice | 2/3PHx | Lipopolysaccharide-resistant depresses replication of DNA and exogenous endotoxin pretreatment stimulates liver regeneration | LPS activates KCs or monocytes to release cytokines such as IL-1 and TNF-α↓ | (58) |
| Innate immune response | Rat | 2/3PHx | Loss of OPN impairs hepatic recruitment of KCs and delays hepatocyte proliferation | LPS levels in the serum↓; IL-6/STAT3 expression ↓ | (63) |
| Innate immune response | Mice | 2/3PHx | FHL2 deficiency exhibits diminished liver regeneration | KCs produces LPS-induced cytokine; Inhibits |
(64) |
| Hemostasis activation | Mice | 2/3PHx | Depleted platelet reduces of hepatocellular proliferation | Hepatic expression and release of pro-inflammatory mediators; Platelet‐derived serotonin↓ | (76) |
| Hemostasis activation | Mice | 2/3PHx | Serotonin promotes regeneration and injury repair | Axis of serotonin -pErk-YAP↑ | (77) |
NO, nitric oxide; ALT, alanine aminotransferase; HSP70, heat shock protein 70; Nrp1, neuropilin1; EGFR, epidermal growth factor receptor; uPA, urokinase-type plasminogen activator; uPAR, urokinase-type plasminogen activator receptor; HGF, hepatocyte growth factor; LPS, lipopolysaccharide; KCs, Kupffer cells; IL-1, interleukin-1; OPN, osteopontin; TNF-α, tumor necrosis factor α; FHL2, four-and-a-half LIM-domain protein 2; NF-κB, nuclear factor kappa B; IL-6, interleukin-6.
Molecular basis of liver regeneration after PHx
The liver regeneration process can generally be divided into three stages: initiation, proliferation and termination, with various molecules participating in the different stages (54). With more extensive research into the molecular mechanism, the search for targeted drugs of liver regeneration has become a particular research focus. In the following, we will review the mechanism of liver regeneration through cytokines, growth factors, and signaling pathways (Figure 2).
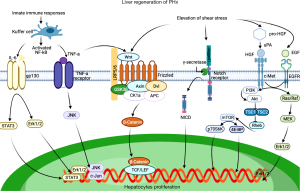
Cytokines
IL-6
Inflammation is a very complex biological response featured by the recruitment, activation, and growth of immune and inflammatory cells, which reduces infection, eliminates damaged cells, and initiates tissue repair and regeneration processes (79).
Inflammatory cells trigger liver regeneration through released cytokines and growth factors. At present, IL-6 and TNF-α are the most widely studied pro-inflammatory cytokines.
When the liver suffers an acute injury, IL-6 plays a critical role in promoting hepatocyte homeostasis and mitosis (80). This means that IL-6 can not only protect the function of the liver, but also promote liver regeneration. During hepatocyte damage, KCs are stimulated, which activates NF-kB and transfers it from the cytoplasm to the nucleus (81). Activated NF-kB then causes KCs to secrete more IL-6 and TNF-α; meanwhile, TNF-α also leads to a massive expression of IL-6 in an autocrine manner (82). IL-6 by binding to the IL-6 receptor (IL-6R) forms the IL-6/IL-6R complex and the complex activates a receptor protein called glycoprotein 130 kDa (gp130), which activates the two pathways: the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway (83,84) and the MEK/Erk pathway. It is known that Jak/Stat is a vital transducing signal related to growth regulation, survival, differentiation, and resistance to pathogens (85). MEK/Erk also plays a significant role in proliferation (86). IL-6 knockout mice have demonstrated alterations in the apoptotic pathways, with a decrease in anti-apoptotic factors (87).
TNF-α
Highly pleiotropic TNF-α induces multiple biological effects such as metabolic activation, necrotic cell death, inflammatory responses, and proliferation (88). On the one hand, TNF-α can activate the anti-apoptotic and pro-inflammatory NF-kB pathway anti-apoptotic by binding to its receptor TNF-R1 (88). On the other hand, TNF activates c-Jun N-terminal kinase (JNK) 1/2 in the liver and subsequently induces the transcription factor c-Jun and its target genes. Since the network of JNK interactions is very complex and not yet fully understood, current studies indicate that it may be involved in triggering hepatocyte proliferation or inducing apoptosis (89). Also, in a culture of hepatocytes, researchers found that TNF-α may participate in remodeling and regeneration of the liver by declining expression of metalloproteinase 9 (MMP9) (90). TNF-α promotes the proliferation of hepatocytes in vivo, and can also play the same role in vitro. Peng et al. found that medium supplemented with TNF-α can make hepatocytes proliferate and undergo continuous passage and culture time for more than 6 months (91). IL-6 and TNF-α are both vital at the beginning of the liver regeneration, and their roles cannot be substituted for each other.
Growth factors
HGF
Blood-derived HGF was first recognized as a mitogen for mature hepatocytes (92). HGF is synthesized in the form of pro-HGF and stored in the liver biomatrix (93). After liver injury or partial hepatectomy, HSCs release pro-HGF, and then release HGF under the cut of uPA and plasminogen protease. HGF binds to c-Met receptors on liver cells to activate downstream signaling pathways, triggering the proliferation and mobilization of liver cells (46,94). The activation of HGF was weakened in uPA-deficient mice after liver injury (49). HGF/c-Met is a key factor for liver growth and function, and contains an intracellular tyrosine kinase domain (95,96). Upon c-Met dimerization, activated kinase facilitates auto-phosphorylation of tyrosine and the downstream signaling pathways are stimulated to produce a verity of biological effects, including proliferation, survival, and angiogenesis (47,48). Once PI3K/Akt/ mammalian target of rapamycin (mTOR) and (Ras or Raf)/MEK/Erk1/2 pathways are blocked, the amplification of hepatocytes is suppressed (48,80,97).
Endothelial growth factor (EGF)
Duodenal Brunner’s glands and salivary glands can synthesize and secrete EGF and KCs product heparin-bound EGF (HB-EGF), which respectively work on hepatocytes via endocrine, paracrine, and autocrine methods (98,99). ErbB family members include ErbB1, ErbB2, ErbB3, and ErbB4. ErbB4 is not expressed in the human liver (100). Even if they have the same receptor, the ErbB ligands exert different effects on liver regeneration and biological responses. In HB-EGF knockout mice, liver regeneration is deficient and decreased because of the delay of hepatocyte DNA synthesis after 1/3 PHx (101). Unlike HGF signaling through c-Met homodimer, EGF signaling via ErbB1, ErbB1 can homo-/hetero-dimerize with ErbB2 or ErbB3, thereby stimulating Ras-Raf-ERK and PI3K-AKT signaling pathways facilitates cell growth, adhesion, and migration (51,52). When EGF is inhibited, animals suffering from hepatectomy experience a delay in hepatocyte division (102).
Transforming growth factor beta (TGF-β)
Although TGF-β is the most well-known signal for the termination of liver regeneration, we have an inadequate understanding of the role of TGF-β in liver regeneration. TGF-β is usually synthesized and secreted in a paracrine way by mesenchymal cells, such as HSCs, KCs, and platelets (103). One hour after PHx, the content of TGF-β significantly improves in the blood. Upon TGF-β originally adhered to the cell membrane surface through decorin (104), the increase in blood content is the result of TGF-β detaching from the membrane surface into the blood and binding to the alpha2-macroglobulin in the blood (105). This process of dissociation from the liver parenchymal cell membrane surface and immobilization in the plasma may be avoided by TGF-β inhibition of the proliferation of liver cells in the early stage (106). In the middle of proliferation, the indirect hepatocyte inhibitor cation-independent mannose 6-phosphate receptor (CIMPR) is expressed, which converts the TGF-β precursor into activated TGF-β and regulates the binding of activated TGF-β to the TGFR (107,108). In addition to the activation of TGF-β itself, the expression and activation of its receptors also has a critical role in activating the entire pathway, and may even be more decisive (109). Studies have found that in the later stage of liver regeneration, the expression of TGFR is significantly increased, which increases the sensitivity of cells to TGF-β. After TGF-β binding to the receptor, the R-Smad protein is phosphorylated and translocated into the nucleus, which activates the transcription of cell cycle inhibitors such as cyclin dependent kinase (CDK) inhibitors, and inhibits cell cycle promoters, such as CDK2/4, Cyclin D/E, etc. These products cause the cell cycle to be blocked (107,108).
Signaling pathways
Wnt/β-Catenin signaling
The rapid activation of Wnt/ β-Catenin is one of the most significant phenomena in the early stage after liver damage. Wnt, as a glycoprotein, is mainly secreted by hepatic non-parenchymal cells (such as KCs and endothelial cells) for β-Catenin activation during regeneration (110,111). Beginning at 5 minutes after hepatectomy, β-Catenin is transiently up-regulated and rapidly transferred to the nucleus, and this process can be maintained for approximately 6 hours (13). The increase of β-Catenin in the nucleus induces the activation of its target genes including Cyclin D1, and transient expression of Cyclin D1 can promote hepatocyte proliferation and regeneration (112,113). The activation of β-Catenin requires the Wnt protein outside the cell to adsorb the destruction complex of β-Catenin to the plasma membrane through its receptor Frizzled and low-density lipoprotein receptor-related protein 5/6 (LRP5/6) to inactivate the degradation function (114). The raising of Wnt may be related to the activation of TNF-α, which can promote the expression of Wnt in KCs. The secretion of Wnt prevents cytoplasmic accumulation of β-Catenin from degrading and entering the nucleolus to activate proliferation (114). β-catenin knockout mice and those >2 months have a pronounced reduction in the hepatic weight ratio (15–25%), and the overexpression of β-catenin directly demonstrates the enhanced liver growth (115-117).
Notch signaling
In mammals, four Notch receptors (Notch1–4) and two types of ligands (Jagged1–2 and DLL1, DLL3–4) have been confirmed. Of these, Notch1 is primarily expressed in hepatocytes and mainly influences the regulation of cell proliferation (118). After Notch1 binds to the Jagged1 ligand, the Notch signaling pathway can be activated. The Notch receptor is then cleaved by the γ-secretase complex, and the Notch1 intracellular domain (NICD1) is released into the cell. NICD1, as the active form of Notch1, can translocate into the nucleus and cause the transcription of Notch target genes related to cell proliferation such as Hes1/5, c-myc, and Cyclin D1 (119-121). However, the mechanism through which Jagged1 is activated quickly in response to damage changes remains unclear. As an important Notch ligand in the liver, Jagged1 is expressed not only in HPCs and cholangiocytes, but in portal vein mesenchymal smooth muscle cells (122,123). The specific knockout of Jagged1 in these cells will lead to abnormal liver development. This indicates that the activation of Jagged1 may be the result of increased portal blood pressure or blood flow rate.
mTOR signaling
mTOR, as a protein kinase, is an important signal molecule in the Akt/tuberous sclerosis complex1/2 (TSC1/2)/mTOR signaling pathway and participates in the regulation of multiple cellular events such as metabolism, cell proliferation, and autophagy. It is activated by the phosphorylation of PI3K/Akt and activated Akt phosphorylates the TSC1/TSC2 complex. The function of the phosphorylated TSC1/TSC2 complex is subsequently inhibited, releasing its inhibition of the small guanosine triphosphatase (GTPase) Rheb, so that Rheb is activated, and the activated form of Rheb positively regulates mTOR (124). It then promotes protein synthesis and cell growth by mediating important downstream signaling molecules, p70 S6 kinase 1 (p70S6K1) and the eukaryotic initiation factor 4E-binding proteins (4E-BPs) (125-127). According to an open experiment, stimulating the mTOR signaling pathway can promote liver growth in Zebrafish and the proliferation of human hepatocytes (128).
Hippo signaling
At present, the activation of liver regeneration signaling is well understood. However, the mechanism through which the body feels the hepatic weight and size recovery remains unclear. Research has shown that the Hippo signaling pathway has a significant role in control the of organ size and tumorigenesis (129,130). Following activation of the Hippo signaling pathway, the mammalian Ste20-like kinases 1/2 (Mst1/2) is triggered and the large tumor suppressor 1/2 (Last1/2) is subsequently activated by phosphorylation. Last1/2 acts as a phosphokinase, which then phosphorylates the Yes-associated protein (YAP) and the transcriptional co-activator with the postsynaptic density protein-95/disks large/zonula occludens-1 [PDZ]-binding motif (TAZ), so that they bind to 14-3-3 and remain in the cytoplasm. Eventually YAP/TAZ is degraded by the ubiquitination pathway and loses its activity.
Upon inactivation of Hippo, the unphosphorylated YAP/TAZ enters the nucleus and binds to the transcription factor TEA-domain-containing proteins (TEADs) to promote cell proliferation (131,132). The inactivation of Mst1/2 and Last1/2, activation of YAP in the initial regeneration stage, and the reactivation of Mst1/2 and Last1/2 following recovery of the liver in terminal regeneration stage confirms the effect of the Hippo signaling pathway in the liver regeneration (53). Meanwhile, the acute inactivation of Hippo signaling can dedifferentiate mature hepatocytes into cells bearing progenitor characteristics (133).
Conclusions
The process of liver regeneration is complex and intricate, consisting of various and interactive pathways. There is sufficient evidence to demonstrate that liver regeneration is similar between humans and rodents. The PHx-induced liver regeneration model is still the first choice for most studies, but it is more common clinically that liver regeneration occurs after injury caused by poisons, alcohol, or immunity. In this case, it is undoubtedly that the liver regeneration model induced by chemical damage is more suitable. In addition to considering the etiologies, different course of the disease will also interfere with model selection. Compared with the PHx-induced liver regeneration model, the liver regeneration process induced by chemical damage is slower. Therefore, it is very important to fully grasp the features of each model and choose a suitable liver regeneration model. Many cytokines, growth factors and signal pathways are interrelated and affect each other. As the understanding of the signal molecules involved in liver regeneration becomes deeper and deeper, we look forward to developing corresponding targeted drugs to improve the efficiency of liver regeneration. Nevertheless, we still do not fully command the mechanism of liver regeneration and increasing the mastery of this knowledge can not only further guide clinical research on how to promote liver regeneration after surgical resection and liver transplantation, but also offer possible targets to treat the dysregulation of regeneration that occurs in chronic hepatic diseases and tumors. Current research work includes the use of animal models as vectors for studying human hepatocytes in vivo, which has shown the uniqueness and importance of the field of liver regeneration.
Acknowledgments
Funding: This study was supported by the 1.3.5 Project for disciplines of excellence, West China Hospital, Sichuan University (ZYGD20009).
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-5234
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-5234). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Michalopoulos GK, DeFrances MC. Liver regeneration. Science 1997;276:60-6. [Crossref] [PubMed]
- Am Esch JS, Schmelzle M, Fürst G, et al. Infusion of CD133+ bone marrow-derived stem cells after selective portal vein embolization enhances functional hepatic reserves after extended right hepatectomy: a retrospective single-center study. Ann Surg 2012;255:79-85. [Crossref] [PubMed]
- Huebert RC, Rakela J. Cellular therapy for liver disease. Mayo Clin Proc 2014;89:414-24. [Crossref] [PubMed]
- Spahr L, Lambert JF, Rubbia-Brandt L, et al. Granulocyte-colony stimulating factor induces proliferation of hepatic progenitors in alcoholic steatohepatitis: a randomized trial. Hepatology 2008;48:221-9. [Crossref] [PubMed]
- Forbes SJ, Newsome PN. Liver regeneration - mechanisms and models to clinical application. Nat Rev Gastroenterol Hepatol 2016;13:473-85. [Crossref] [PubMed]
- Meier M, Andersen KJ, Knudsen AR, et al. Liver regeneration is dependent on the extent of hepatectomy. J Surg Res 2016;205:76-84. [Crossref] [PubMed]
- Higgins GM, Anderson RM. Experimental pathology of the liver. I. Restoration of the liver of the white rat following partial surgical removal. Arch Pathol 1931;12:1186-202.
- Yagi S, Hirata M, Miyachi Y, et al. Liver Regeneration after Hepatectomy and Partial Liver Transplantation. Int J Mol Sci 2020;21:8414. [Crossref] [PubMed]
- Miyaoka Y, Ebato K, Kato H, et al. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol 2012;22:1166-75. [Crossref] [PubMed]
- Fausto N. Liver regeneration: from laboratory to clinic. Liver Transpl 2001;7:835-44. [Crossref] [PubMed]
- Bhave VS, Mars W, Donthamsetty S, et al. Regulation of liver growth by glypican 3, CD81, hedgehog, and Hhex. Am J Pathol 2013;183:153-9. [Crossref] [PubMed]
- Michalopoulos GK. Advances in liver regeneration. Expert Rev Gastroenterol Hepatol 2014;8:897-907. [Crossref] [PubMed]
- Monga SP, Pediaditakis P, Mule K, et al. Changes in WNT/beta-catenin pathway during regulated growth in rat liver regeneration. Hepatology 2001;33:1098-109. [Crossref] [PubMed]
- Liu X, Xu J, Rosenthal S, et al. Identification of Lineage-Specific Transcription Factors That Prevent Activation of Hepatic Stellate Cells and Promote Fibrosis Resolution. Gastroenterology 2020;158:1728-1744.e14. [Crossref] [PubMed]
- Issa R, Zhou X, Trim N, et al. Mutation in collagen-1 that confers resistance to the action of collagenase results in failure of recovery from CCl4-induced liver fibrosis, persistence of activated hepatic stellate cells, and diminished hepatocyte regeneration. FASEB J 2003;17:47-9. [Crossref] [PubMed]
- Iredale JP. Models of liver fibrosis: exploring the dynamic nature of inflammation and repair in a solid organ. J Clin Invest 2007;117:539-48. [Crossref] [PubMed]
- Duffield JS, Forbes SJ, Constandinou CM, et al. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J Clin Invest 2005;115:56-65. [Crossref] [PubMed]
- Feng L, Cai L, He GL, et al. Novel D-galactosamine-induced cynomolgus monkey model of acute liver failure. World J Gastroenterol 2017;23:7572-83. [Crossref] [PubMed]
- Hishinuma I, Nagakawa J, Hirota K, et al. Involvement of tumor necrosis factor-alpha in development of hepatic injury in galactosamine-sensitized mice. Hepatology 1990;12:1187-91. [Crossref] [PubMed]
- Wang CC, Cheng PY, Peng YJ, et al. Naltrexone protects against lipopolysaccharide/D-galactosamine-induced hepatitis in mice. J Pharmacol Sci 2008;108:239-47. [Crossref] [PubMed]
- Nishiguchi S, Kuroki T, Takeda T, et al. Effects of putrescine on D-galactosamine-induced acute liver failure in rats. Hepatology 1990;12:348-53. [Crossref] [PubMed]
- Nakama T, Hirono S, Moriuchi A, et al. Etoposide prevents apoptosis in mouse liver with D-galactosamine/lipopolysaccharide-induced fulminant hepatic failure resulting in reduction of lethality. Hepatology 2001;33:1441-50. [Crossref] [PubMed]
- Gehrke N, Hövelmeyer N, Waisman A, et al. Hepatocyte-specific deletion of IL1-RI attenuates liver injury by blocking IL-1 driven autoinflammation. J Hepatol 2018;68:986-95. [Crossref] [PubMed]
- Chung H, Kim HJ, Jang KS, et al. Comprehensive analysis of differential gene expression profiles on D-galactosamine-induced acute mouse liver injury and regeneration. Toxicology 2006;227:136-44. [Crossref] [PubMed]
- Bernal W, Lee WM, Wendon J, et al. Acute liver failure: A curable disease by 2024? J Hepatol 2015;62:S112-20. [Crossref] [PubMed]
- Jaeschke H, McGill MR, Ramachandran A. Oxidant stress, mitochondria, and cell death mechanisms in drug-induced liver injury: lessons learned from acetaminophen hepatotoxicity. Drug Metab Rev 2012;44:88-106. [Crossref] [PubMed]
- Bhushan B, Apte U. Liver Regeneration after Acetaminophen Hepatotoxicity: Mechanisms and Therapeutic Opportunities. Am J Pathol 2019;189:719-29. [Crossref] [PubMed]
- Ni HM, McGill MR, Chao X, et al. Removal of acetaminophen protein adducts by autophagy protects against acetaminophen-induced liver injury in mice. J Hepatol 2016;65:354-62. [Crossref] [PubMed]
- Yan M, Ye L, Yin S, et al. Glycycoumarin protects mice against acetaminophen-induced liver injury predominantly via activating sustained autophagy. Br J Pharmacol 2018;175:3747-57. [Crossref] [PubMed]
- Mohar I, Stamper BD, Rademacher PM, et al. Acetaminophen-induced liver damage in mice is associated with gender-specific adduction of peroxiredoxin-6. Redox Biol 2014;2:377-87. [Crossref] [PubMed]
- Díaz-Guerra MJ, Velasco M, Martín-Sanz P, et al. Nuclear factor kappaB is required for the transcriptional control of type II NO synthase in regenerating liver. Biochem J 1997;326:791-7. [Crossref] [PubMed]
- Díez-Fernández C, Boscá L, Fernández-Simón L, et al. Relationship between genomic DNA ploidy and parameters of liver damage during necrosis and regeneration induced by thioacetamide. Hepatology 1993;18:912-8. [Crossref] [PubMed]
- Fernández-Martínez A, Callejas NA, Casado M, et al. Thioacetamide-induced liver regeneration involves the expression of cyclooxygenase 2 and nitric oxide synthase 2 in hepatocytes. J Hepatol 2004;40:963-70. [Crossref] [PubMed]
- Grompe M. Fah Knockout Animals as Models for Therapeutic Liver Repopulation. Adv Exp Med Biol 2017;959:215-30. [Crossref] [PubMed]
- Grompe M, Strom S. Mice with human livers. Gastroenterology 2013;145:1209-14. [Crossref] [PubMed]
- Sato Y, Koyama S, Tsukada K, et al. Acute portal hypertension reflecting shear stress as a trigger of liver regeneration following partial hepatectomy. Surg Today 1997;27:518-26. [Crossref] [PubMed]
- Sato Y, Tsukada K, Hatakeyama K. Role of shear stress and immune responses in liver regeneration after a partial hepatectomy. Surg Today 1999;29:1-9. [Crossref] [PubMed]
- Fan YD, Praet M, Van Huysse J, et al. Effects of portal vein arterialization on liver regeneration after partial hepatectomy in the rat. Liver Transpl 2002;8:146-52. [Crossref] [PubMed]
- Marubashi S, Sakon M, Nagano H, et al. Effect of portal hemodynamics on liver regeneration studied in a novel portohepatic shunt rat model. Surgery 2004;136:1028-37. [Crossref] [PubMed]
- Schoen JM, Wang HH, Minuk GY, et al. Shear stress-induced nitric oxide release triggers the liver regeneration cascade. Nitric Oxide 2001;5:453-64. [Crossref] [PubMed]
- Carnovale CE, Ronco MT. Role of nitric oxide in liver regeneration. Ann Hepatol 2012;11:636-47. [Crossref] [PubMed]
- Kumamoto T, Togo S, Ishibe A, et al. Role of nitric oxide synthesized by nitric oxide synthase 2 in liver regeneration. Liver Int 2008;28:865-77. [Crossref] [PubMed]
- Fausto N, Campbell JS, Riehle KJ. Liver regeneration. J Hepatol 2012;57:692-4. [Crossref] [PubMed]
- Kawai M, Naruse K, Komatsu S, et al. Mechanical stress-dependent secretion of interleukin 6 by endothelial cells after portal vein embolization: clinical and experimental studies. J Hepatol 2002;37:240-6. [Crossref] [PubMed]
- Sokabe T, Yamamoto K, Ohura N, et al. Differential regulation of urokinase-type plasminogen activator expression by fluid shear stress in human coronary artery endothelial cells. Am J Physiol Heart Circ Physiol 2004;287:H2027-34. [Crossref] [PubMed]
- Mars WM, Liu ML, Kitson RP, et al. Immediate early detection of urokinase receptor after partial hepatectomy and its implications for initiation of liver regeneration. Hepatology 1995;21:1695-701. [PubMed]
- Ueki T, Kaneda Y, Tsutsui H, et al. Hepatocyte growth factor gene therapy of liver cirrhosis in rats. Nat Med 1999;5:226-30. [Crossref] [PubMed]
- Fajardo-Puerta AB, Mato Prado M, Frampton AE, et al. Gene of the month: HGF. J Clin Pathol 2016;69:575-9. [Crossref] [PubMed]
- Shimizu M, Hara A, Okuno M, et al. Mechanism of retarded liver regeneration in plasminogen activator-deficient mice: impaired activation of hepatocyte growth factor after Fas-mediated massive hepatic apoptosis. Hepatology 2001;33:569-76. [Crossref] [PubMed]
- Stolz DB, Mars WM, Petersen BE, et al. Growth factor signal transduction immediately after two-thirds partial hepatectomy in the rat. Cancer Res 1999;59:3954-60. [PubMed]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2001;2:127-37. [Crossref] [PubMed]
- Carver RS, Stevenson MC, Scheving LA, et al. Diverse expression of ErbB receptor proteins during rat liver development and regeneration. Gastroenterology 2002;123:2017-27. [Crossref] [PubMed]
- Mao SA, Glorioso JM, Nyberg SL. Liver regeneration. Transl Res 2014;163:352-62. [Crossref] [PubMed]
- Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology 2006;43:S45-53. [Crossref] [PubMed]
- El Kasmi KC, Anderson AL, Devereaux MW, et al. Toll-like receptor 4-dependent Kupffer cell activation and liver injury in a novel mouse model of parenteral nutrition and intestinal injury. Hepatology 2012;55:1518-28. [Crossref] [PubMed]
- Mäck C, Jungermann K, Götze O, et al. Anaphylatoxin C5a actions in rat liver: synergistic enhancement by C5a of lipopolysaccharide-dependent alpha(2)-macroglobulin gene expression in hepatocytes via IL-6 release from Kupffer cells. J Immunol 2001;167:3972-9. [Crossref] [PubMed]
- Koleva M, Schlaf G, Landmann R, et al. Induction of anaphylatoxin C5a receptors in rat hepatocytes by lipopolysaccharide in vivo: mediation by interleukin-6 from Kupffer cells. Gastroenterology 2002;122:697-708. [Crossref] [PubMed]
- Cornell RP, Liljequist BL, Bartizal KF. Depressed liver regeneration after partial hepatectomy of germ-free, athymic and lipopolysaccharide-resistant mice. Hepatology 1990;11:916-22. [Crossref] [PubMed]
- Tran-Thi TA, Decker K, Baeuerle PA. Differential activation of transcription factors NF-kappa B and AP-1 in rat liver macrophages. Hepatology 1995;22:613-9. [Crossref] [PubMed]
- He G, Karin M. NF-κB and STAT3 - key players in liver inflammation and cancer. Cell Res 2011;21:159-68. [Crossref] [PubMed]
- Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell 2002;109:S81-96. [Crossref] [PubMed]
- Ozaki M. Cellular and molecular mechanisms of liver regeneration: Proliferation, growth, death and protection of hepatocytes. Semin Cell Dev Biol 2020;100:62-73. [Crossref] [PubMed]
- Wen Y, Feng D, Wu H, et al. Defective Initiation of Liver Regeneration in Osteopontin-Deficient Mice after Partial Hepatectomy due to Insufficient Activation of IL-6/Stat3 Pathway. Int J Biol Sci 2015;11:1236-47. [Crossref] [PubMed]
- Dahan J, Nouët Y, Jouvion G, et al. LIM-only protein FHL2 activates NF-κB signaling in the control of liver regeneration and hepatocarcinogenesis. Mol Cell Biol 2013;33:3299-308. [Crossref] [PubMed]
- Rahbari NN, Garden OJ, Padbury R, et al. Posthepatectomy liver failure: a definition and grading by the International Study Group of Liver Surgery (ISGLS). Surgery 2011;149:713-24. [Crossref] [PubMed]
- Luedde T, Schwabe RF. NF-κB in the liver--linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2011;8:108-18. [Crossref] [PubMed]
- Starlinger P, Luyendyk JP, Groeneveld DJ. Hemostasis and Liver Regeneration. Semin Thromb Hemost 2020;46:735-42. [Crossref] [PubMed]
- Groeneveld D, Pereyra D, Veldhuis Z, et al. Intrahepatic fibrin(ogen) deposition drives liver regeneration after partial hepatectomy in mice and humans. Blood 2019;133:1245-56. [Crossref] [PubMed]
- Tatsumi K, Ohashi K, Taminishi S, et al. Regulation of coagulation factors during liver regeneration in mice: mechanism of factor VIII elevation in plasma. Thromb Res 2011;128:54-61. [Crossref] [PubMed]
- Matsuo R, Nakano Y, Ohkohchi N. Platelet administration via the portal vein promotes liver regeneration in rats after 70% hepatectomy. Ann Surg 2011;253:759-63. [Crossref] [PubMed]
- Han S, Park HW, Song JH, et al. Association Between Intraoperative Platelet Transfusion and Early Graft Regeneration in Living Donor Liver Transplantation. Ann Surg 2016;264:1065-72. [Crossref] [PubMed]
- Matsuo R, Ohkohchi N, Murata S, et al. Platelets Strongly Induce Hepatocyte Proliferation with IGF-1 and HGF In Vitro. J Surg Res 2008;145:279-86. [Crossref] [PubMed]
- Furrer K, Rickenbacher A, Tian Y, et al. Serotonin reverts age-related capillarization and failure of regeneration in the liver through a VEGF-dependent pathway. Proc Natl Acad Sci U S A 2011;108:2945-50. [Crossref] [PubMed]
- Meyer J, Balaphas A, Fontana P, et al. Platelet Interactions with Liver Sinusoidal Endothelial Cells and Hepatic Stellate Cells Lead to Hepatocyte Proliferation. Cells 2020;9:1243. [Crossref] [PubMed]
- Lesurtel M, Graf R, Aleil B, et al. Platelet-derived serotonin mediates liver regeneration. Science 2006;312:104-7. [Crossref] [PubMed]
- Nocito A, Georgiev P, Dahm F, et al. Platelets and platelet-derived serotonin promote tissue repair after normothermic hepatic ischemia in mice. Hepatology 2007;45:369-76. [Crossref] [PubMed]
- Fang Y, Liu C, Shu B, et al. Axis of serotonin -pERK-YAP in liver regeneration. Life Sci 2018;209:490-7. [Crossref] [PubMed]
- Lehwald N, Duhme C, Pinchuk I, et al. Platelets Boost Recruitment of CD133+ Bone Marrow Stem Cells to Endothelium and the Rodent Liver-The Role of P-Selectin/PSGL-1 Interactions. Int J Mol Sci 2020;21:6431. [Crossref] [PubMed]
- Karin M, Clevers H. Reparative inflammation takes charge of tissue regeneration. Nature 2016;529:307-15. [Crossref] [PubMed]
- Fujiyoshi M, Ozaki M. Molecular mechanisms of liver regeneration and protection for treatment of liver dysfunction and diseases. J Hepatobiliary Pancreat Sci 2011;18:13-22. [Crossref] [PubMed]
- Valizadeh A, Majidinia M, Samadi-Kafil H, et al. The roles of signaling pathways in liver repair and regeneration. J Cell Physiol 2019; Epub ahead of print. [Crossref] [PubMed]
- Kishimoto T. IL-6: from its discovery to clinical applications. Int Immunol 2010;22:347-52. [Crossref] [PubMed]
- Kishimoto T. Interleukin-6: discovery of a pleiotropic cytokine. Arthritis Res Ther 2006;8:S2. [Crossref] [PubMed]
- Schmidt-Arras D, Rose-John S. IL-6 pathway in the liver: From physiopathology to therapy. J Hepatol 2016;64:1403-15. [Crossref] [PubMed]
- Dodington DW, Desai HR, Woo M. JAK/STAT - Emerging Players in Metabolism. Trends Endocrinol Metab 2018;29:55-65. [Crossref] [PubMed]
- Degirmenci U, Wang M, Hu J. Targeting Aberrant RAS/RAF/MEK/ERK Signaling for Cancer Therapy. Cells 2020;9:198. [Crossref] [PubMed]
- Kovalovich K, Li W, DeAngelis R, et al. Interleukin-6 protects against Fas-mediated death by establishing a critical level of anti-apoptotic hepatic proteins FLIP, Bcl-2, and Bcl-xL. J Biol Chem 2001;276:26605-13. [Crossref] [PubMed]
- Liedtke C, Trautwein C. The role of TNF and Fas dependent signaling in animal models of inflammatory liver injury and liver cancer. Eur J Cell Biol 2012;91:582-9. [Crossref] [PubMed]
- Lemire JM, Shiojiri N, Fausto N. Oval cell proliferation and the origin of small hepatocytes in liver injury induced by D-galactosamine. Am J Pathol 1991;139:535-52. [PubMed]
- Haruyama T, Ajioka I, Akaike T, et al. Regulation and significance of hepatocyte-derived matrix metalloproteinases in liver remodeling. Biochem Biophys Res Commun 2000;272:681-6. [Crossref] [PubMed]
- Peng WC, Logan CY, Fish M, et al. Inflammatory Cytokine TNFα Promotes the Long-Term Expansion of Primary Hepatocytes in 3D Culture. Cell 2018;175:1607-1619.e15. [Crossref] [PubMed]
- Gohda E, Tsubouchi H, Nakayama H, et al. Purification and partial characterization of hepatocyte growth factor from plasma of a patient with fulminant hepatic failure. J Clin Invest 1988;81:414-9. [Crossref] [PubMed]
- Cienfuegos JA, Rotellar F, Baixauli J, et al. Liver regeneration--the best kept secret. A model of tissue injury response. Rev Esp Enferm Dig 2014;106:171-94. [PubMed]
- Mohammed FF, Khokha R. Thinking outside the cell: proteases regulate hepatocyte division. Trends Cell Biol 2005;15:555-63. [Crossref] [PubMed]
- Naldini L, Vigna E, Narsimhan RP, et al. Hepatocyte growth factor (HGF) stimulates the tyrosine kinase activity of the receptor encoded by the proto-oncogene c-MET. Oncogene 1991;6:501-4. [PubMed]
- Bottaro DP, Rubin JS, Faletto DL, et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 1991;251:802-4. [Crossref] [PubMed]
- Liu JJ, Li Y, Chen WS, et al. Shp2 deletion in hepatocytes suppresses hepatocarcinogenesis driven by oncogenic β-Catenin, PIK3CA and MET. J Hepatol 2018;69:79-88. [Crossref] [PubMed]
- Khan MGM, Ghosh A, Variya B, et al. Hepatocyte growth control by SOCS1 and SOCS3. Cytokine 2019;121:154733 [Crossref] [PubMed]
- Jones DE Jr, Tran-Patterson R, Cui DM, et al. Epidermal growth factor secreted from the salivary gland is necessary for liver regeneration. Am J Physiol 1995;268:G872-8. [PubMed]
- Michalopoulos GK, Khan Z. Liver regeneration, growth factors, and amphiregulin. Gastroenterology 2005;128:503-6. [Crossref] [PubMed]
- Mitchell C, Nivison M, Jackson LF, et al. Heparin-binding epidermal growth factor-like growth factor links hepatocyte priming with cell cycle progression during liver regeneration. J Biol Chem 2005;280:2562-8. [Crossref] [PubMed]
- Natarajan A, Wagner B, Sibilia M. The EGF receptor is required for efficient liver regeneration. Proc Natl Acad Sci U S A 2007;104:17081-6. [Crossref] [PubMed]
- Braun L, Mead JE, Panzica M, et al. Transforming growth factor beta mRNA increases during liver regeneration: a possible paracrine mechanism of growth regulation. Proc Natl Acad Sci U S A 1988;85:1539-43. [Crossref] [PubMed]
- Dudás J, Kovalszky I, Gallai M, et al. Expression of decorin, transforming growth factor-beta 1, tissue inhibitor metalloproteinase 1 and 2, and type IV collagenases in chronic hepatitis. Am J Clin Pathol 2001;115:725-35. [Crossref] [PubMed]
- Webb DJ, Roadcap DW, Dhakephalkar A, et al. A 16-amino acid peptide from human alpha2-macroglobulin binds transforming growth factor-beta and platelet-derived growth factor-BB. Protein Sci 2000;9:1986-92. [Crossref] [PubMed]
- Michalopoulos GK. Principles of liver regeneration and growth homeostasis. Compr Physiol 2013;3:485-513. [Crossref] [PubMed]
- Liu M, Chen P. Proliferation-inhibiting pathways in liver regeneration Mol Med Rep 2017;16:23-35. (Review). [Crossref] [PubMed]
- Hines IN, Kremer M, Isayama F, et al. Impaired liver regeneration and increased oval cell numbers following T cell-mediated hepatitis. Hepatology 2007;46:229-41. [Crossref] [PubMed]
- Nishikawa Y, Wang M, Carr BI. Changes in TGF-beta receptors of rat hepatocytes during primary culture and liver regeneration: increased expression of TGF-beta receptors associated with increased sensitivity to TGF-beta-mediated growth inhibition. J Cell Physiol 1998;176:612-23. [Crossref] [PubMed]
- Yang J, Mowry LE, Nejak-Bowen KN, et al. β-catenin signaling in murine liver zonation and regeneration: a Wnt-Wnt situation! Hepatology 2014;60:964-76. [Crossref] [PubMed]
- Ding BS, Nolan DJ, Butler JM, et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature 2010;468:310-5. [Crossref] [PubMed]
- Nelsen CJ, Rickheim DG, Timchenko NA, et al. Transient expression of cyclin D1 is sufficient to promote hepatocyte replication and liver growth in vivo. Cancer Res 2001;61:8564-8. [PubMed]
- Rickheim DG, Nelsen CJ, Fassett JT, et al. Differential regulation of cyclins D1 and D3 in hepatocyte proliferation. Hepatology 2002;36:30-8. [Crossref] [PubMed]
- Russell JO, Monga SP. Wnt/β-Catenin Signaling in Liver Development, Homeostasis, and Pathobiology. Annu Rev Pathol 2018;13:351-78. [Crossref] [PubMed]
- Tan X, Behari J, Cieply B, et al. Conditional deletion of beta-catenin reveals its role in liver growth and regeneration. Gastroenterology 2006;131:1561-72. [Crossref] [PubMed]
- Sekine S, Lan BY, Bedolli M, et al. Liver-specific loss of beta-catenin blocks glutamine synthesis pathway activity and cytochrome p450 expression in mice. Hepatology 2006;43:817-25. [Crossref] [PubMed]
- Tan X, Apte U, Micsenyi A, et al. Epidermal growth factor receptor: a novel target of the Wnt/beta-catenin pathway in liver. Gastroenterology 2005;129:285-302. [Crossref] [PubMed]
- Morell CM, Strazzabosco M. Notch signaling and new therapeutic options in liver disease. J Hepatol 2014;60:885-90. [Crossref] [PubMed]
- Köhler C, Bell AW, Bowen WC, et al. Expression of Notch-1 and its ligand Jagged-1 in rat liver during liver regeneration. Hepatology 2004;39:1056-65. [Crossref] [PubMed]
- Adams JM, Jafar-Nejad H. The Roles of Notch Signaling in Liver Development and Disease. Biomolecules 2019;9:608. [Crossref] [PubMed]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 2009;137:216-33. [Crossref] [PubMed]
- Hofmann JJ, Zovein AC, Koh H, et al. Jagged1 in the portal vein mesenchyme regulates intrahepatic bile duct development: insights into Alagille syndrome. Development 2010;137:4061-72. [Crossref] [PubMed]
- Kodama Y, Hijikata M, Kageyama R, et al. The role of notch signaling in the development of intrahepatic bile ducts. Gastroenterology 2004;127:1775-86. [Crossref] [PubMed]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012;149:274-93. [Crossref] [PubMed]
- Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 2020;21:183-203. [Crossref] [PubMed]
- Fingar DC, Salama S, Tsou C, et al. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev 2002;16:1472-87. [Crossref] [PubMed]
- Espeillac C, Mitchell C, Celton-Morizur S, et al. S6 kinase 1 is required for rapamycin-sensitive liver proliferation after mouse hepatectomy. J Clin Invest 2011;121:2821-32. [Crossref] [PubMed]
- Chaturantabut S, Shwartz A, Evason KJ, et al. Estrogen Activation of G-Protein-Coupled Estrogen Receptor 1 Regulates Phosphoinositide 3-Kinase and mTOR Signaling to Promote Liver Growth in Zebrafish and Proliferation of Human Hepatocytes. Gastroenterology 2019;156:1788-1804.e13. [Crossref] [PubMed]
- Hong L, Cai Y, Jiang M, et al. The Hippo signaling pathway in liver regeneration and tumorigenesis. Acta Biochim Biophys Sin (Shanghai) 2015;47:46-52. [Crossref] [PubMed]
- Patel SH, Camargo FD, Yimlamai D. Hippo Signaling in the Liver Regulates Organ Size, Cell Fate, and Carcinogenesis. Gastroenterology 2017;152:533-45. [Crossref] [PubMed]
- Moya IM, Halder G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat Rev Mol Cell Biol 2019;20:211-26. [Crossref] [PubMed]
- Yimlamai D, Fowl BH, Camargo FD. Emerging evidence on the role of the Hippo/YAP pathway in liver physiology and cancer. J Hepatol 2015;63:1491-501. [Crossref] [PubMed]
- Yimlamai D, Christodoulou C, Galli GG, et al. Hippo pathway activity influences liver cell fate. Cell 2014;157:1324-38. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


