
Quantitative immunohistochemistry (IHC) analysis of biomarker combinations for human esophageal squamous cell carcinoma
Introduction
Esophageal carcinoma is one of the most common cancers and causes more than 400,000 deaths annually worldwide (1,2). Esophageal cancer is classified by histology as esophageal squamous carcinoma (ESCC) and esophageal adenocarcinoma. ESCC is the most common histological subtype and represents up to 90% of cases in the highest risk areas of the “esophageal cancer belt,” which stretches from Northern Iran through Central Asia to China (1,3-5). Currently, there are few tumor biomarkers for ESCC. Such biomarkers are important for early diagnosis, staging, prognosis, and guiding the development of effective targeted therapies. To identify new biomarkers specific to ESCC, this study evaluated the expression of the following 4 conventional immunostaining proteins in cancer diagnosis: proliferating cell nuclear antigen (PCNA), tumor suppressor protein p53, epidermal growth factor receptor (EGFR), and vascular endothelial growth factor (VEGF).
PCNA acts as a molecular coordinator in maintaining genomic integrity at both the genetic and epigenetic levels (6,7). PCNA is a marker of esophageal cellular proliferation. It can clearly demarcate proliferating areas (8). Some research findings have supported the hypothesis that PCNA targeting may be a way to specifically target cancerous cells by comparing the sensitivity of cancer and non-transformed cell lines in cell cultures (9-11). Tumor protein p53 is a well-known tumor suppressor. It is encoded by gene TP53, which is the most frequently mutated gene (>50%) in human cancer (12). Somatic alterations of TP53 were found in a large proportion (63–93%) of esophageal cancer cases (13-15). Protein p53 is a promising candidate biomarker for predicting ESCC patients’ responses to chemotherapy (16). The EGFR is a receptor for members of the epidermal growth factor family of extracellular protein ligands. The over-expression of EGFR is thought to be related to mutations of the EGFR gene, which lead to the constant activation of EGFR and uncontrolled cell division (17). The mutation of the EGFR gene is associated with the development of a wide spectrum of tumors, including epithelial tumors of the head and neck (18), squamous cell carcinoma of the lung, and anal cancers (19). VEGF is a signal protein that stimulates the development of vessels (20). VEGF activity has some effects on reinforcing microvascular permeability and stimulating monocyte/macrophage migration (21). Some previous studies have reported on these proteins in relation to cancers other than ESCC (11,12,17). Some previous studies have identified molecular biomarkers of ESCC (22,23). In the present study, we systematically evaluated the expression of 4 proteins for ESCC with clinical samples. We found an IHC panel from these biomarkers which existing pathology services could easily adopt as a routine test to determine the status of these three molecular biomarkers.
The IHC pattern and intensity of tumor biomarkers of ESCC accordingly reflect the Tumor Mutational Burden (TMB) and neoantigen load in some extent (24). TMB has emerged to predict patients’ respond to immunotherapy. Some clinical trials have shown that high TMB increases the efficacy of immune checkpoint blockades in cancer immunotherapy (25). While simultaneous radiotherapy and chemotherapy for ESCC may decrease TMB in ESCC cells. IHC analyzes being tested starting before not after concurrent chemoradiation therapy would truly reflect the characteristics of the tumor and to identify patients most likely to respond to immunotherapy. In our study, we evaluated the expression of the biomarkers specific to ESCC by samples without preoperative anti-tumor therapy, which might help in ESCC immunotherapy.
We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/atm-21-2950).
Methods
Sample source and collection
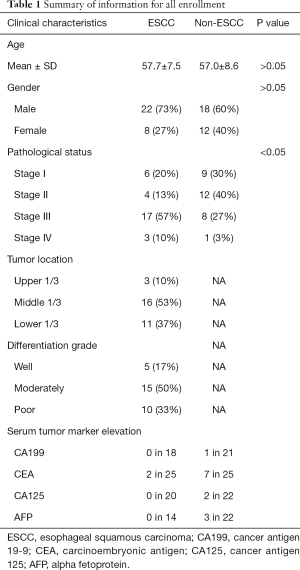
We examined tissue samples from 30 primary ESCC patients and randomly selected tissue samples from 30 patients with tumors (other than tumors of the esophagus) as the control group. All patients underwent curative surgery without preoperative anti-tumor therapy between June 2005 and January 2018 at The Fifth Affiliated Hospital, Sun Yat-sen University, Zhuhai, Guangdong Province, China. To conceal patients’ health data and protect sensitive information, the personal information of all patients was masked in accordance with the standard electronic health record (EHR) process (a common technique used to alter information in a patient’s EHR that includes data encryption, obfuscation, hashing, exclusion, and perturbation). Patients with undifferentiated carcinomas or other malignant tumors of the esophagus were excluded from the study. In relation to the ESCC cases, tumor, nodes, and metastases (TNM) stage and lymph node status were classified according to the American Joint Committee on Cancer’s (AJCC’s) system (8th edition). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional research ethics board of the Fifth Affiliated Hospital, Sun Yat-sen university (No. 2019 K20-1). Because of the retrospective nature of the study, patient consent for inclusion was waived.
Immunohistochemistry (IHC)
The IHC procedure was performed according to a manual [see our previous publications (26,27)]. All surgical pathology specimens from the thoracic ESCCs of 30 patients and tumors of 30 non-ESCC patients were formalin-fixed. The tissue samples were paraffin-embedded and sectioned at 4-µm thickness. The slides were antigen retrieved with citrate buffer (pH 6.0) by heating in an autoclave. The slides were washed with phosphate-buffered saline (PBS), and endogenous peroxidase was quenched and blocked for 10 min at room temperature. The slides were incubated for 1 hour at 37 °C with specific primary antibodies (rabbit anti-human EGFR monoclonal antibody: cat. No. RMA-0554; clone EP38Y; mouse anti-human VEGF monoclonal antibody: cat. No. MAB-0243; clone VG1; mouse anti-human PCNA monoclonal antibody: cat. No MAB-0145. clone PC10; mouse anti-human p53 monoclonal antibody: cat. No. MAB-0674, clone MX008). All of the primary antibodies were purchased from MXB Biotechnologies, Fuzhou, China. Next, the slides were washed 3 times with PBS, and secondary antibodies [sheep anti-mouse immunoglobulin G (IgG) polymer and sheep anti-rabbit IgG polymer, purchased from MXB Biotechnologies, Fuzhou, China] were incubated for 30 minutes at room temperature. The slides were then washed 3 times with PBS, and 3,3'-diaminobenzidine tetrahydrochloride (DAB) was applied to detect the presence of each biomarker in the clinical samples. Finally, the slides were counterstained with hematoxylin for nucleic staining.
Quantification of IHC
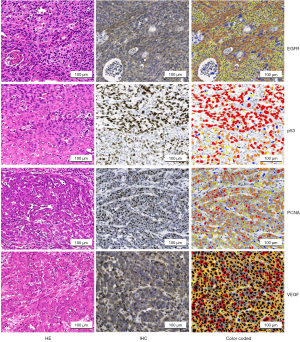
All stained slides were digitally scanned at 40× or 20× with a Pannoramic scan (3DHistech, Budapest, Hungary). To minimize inter-observer variations, IHC staining was evaluated by a computer-assisted diagnosis application (CaseViewer version:2.2). The digital slides were visualized and analyzed with QuantCenter (3DHistech, Budapest, Hungary). The area of each tumor was identified manually for each digital slide. Five different regions of tumor area were outlined; each region contained more than 800 cells. The number of stained cells in each region was calculated with corresponding quantification modules (nuclear quant for PCNA and p53, membrane quant for EGFR, and cell quant for VEGF). The modules reported negative and positive cell counts and their percentages, as stratified by the intensity of staining (negative, weak, intermediate, or strong) (see Figure 1). Finally, the H-score for each region was calculated at a default setting, which was determined by the percentage of positive cells and their intensity (H-score = Σpi(i+1) where “pi” represents the percentage of positive cell counts in total cell counts, and “i” represents the intensity) (28,29). To evaluate the accuracy of the computer-assisted measurement, the computerized images and the computer-assisted measurements were verified with at least 2 professional pathologist-based scoring results.
Western blot
Fresh tissue samples from the ESCC tumor and adjacent normal tissue were retrieved from five patients. Total protein was extracted by lysis buffer (RIPA buffer, thermo, 89900, add 1% Triton X 100, and 1% protease inhibitor cocktail before use), resolved by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride membranes. The membranes were blocked in Tris-buffered saline with Tween-20(0.02%) solution (TBST) containing 5% slim milk and probed with primary antibodies [an anti-PCNA antibody from MXB Biotechnologies, MAB-0145, an anti-EGFR antibody from MXB Biotechnologies, RMA-0554, and a beta actin antibody from ThermoFisher (MA5-15739)] overnight at 4 °C. They were then washed and incubated with a secondary antibody conjugated with horseradish peroxidase. β-actin protein levels were used as a loading control. For detection, the membranes were incubated with Supersignal West Pico PLUS Chemiluminescent Substrate (Thermo) for 5 min and then exposed by the ChemiDoc XRS+ imaging System (BioRad). The intensity of the protein bands was quantified using image Lab (BioRad).
Statistics and receiver operating characteristic analysis
The statistical analysis was performed using SPSS Statistics version 20.0 software (SPSS, Chicago, IL, USA), Graphpad Prism version 6.01 software (GraphPad Software, San Diego, USA), and MedCalc 18.2.1 software (MedCalc Software, Ostend, Belgium). A statistically significant difference was defined as P<0.05. Descriptive statistics were used to summarize patients’ clinical features. We compared individual assays between the ESCC and non-ESCC groups using an independent-samples Mann-Whitney U test. The student's t-test was used to calculate the statistical significance of the levels of EGFR and PCNA in a western blot. The diagnosis performance of each marker was evaluated using a receiver operating characteristic (ROC) curve, and the area under the curve (AUC) value derived from each curve was estimated (as previously described) and compared using Delong’s test (30). To simplify the IHC results, the optimal cut-off points for the low or high expression for each marker were determined by the ROC. The optimal cut-off point was identified as the point corresponding to the optimal sensitivity and specificity. The optimal sensitivity and specificity values were defined as the values yielding the minimal value for (1−sensitivity)2 + (1−specificity)2 (31). We estimated sensitivity, specificity, and predictive values for PCNA/p53/EGFR/VEGF. All confidence intervals (CIs) were reported as 2-sided binomial distributions at 95% confidence.
Results
Expression quantification in IHC
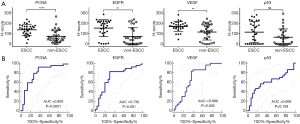
We compared EGFR, p53, PCNA, and VEGF IHC expression between the ESCC and non-ESCC groups for which age and gender had been matched. This sample set comprised individual tumor samples from 30 ESCC and 30 non-ESCC patients. Table 1 provides a summary of the clinical pathological features of patients. The ESCC cases were distributed across the AJCC stages of I-IV. The non-ESCC cases included breast cancer (n=8), liver cancer (n=2), non-small cell lung cancer (n=5), colorectal cancer (n=10), gastric adenocarcinoma (n=4), and renal carcinoma (n=1) cases. There was no significant difference between the ESCC and non-ESCC groups in terms of gender and age distribution (P=0.41 and P=0.73, respectively). In total, 240 slices were included in this study. For each tumoral section in these slices, the immune stain features of 4–6 views, which contained 6,250–41,100 cells in hot spots, were evaluated. The expression of the 4 proteins in the ESCC resection tumor specimens were relatively higher than those of the non-ESCC specimens based on quantitative H-scores. The H-scores analysis indicated that EGFR (the median of ESCC vs. non-ESCC, 177.0 vs. 47.08; P=0.0014), PCNA (146.5 vs. 52.32; P<0.0001), and VEGF (177.1 vs. 128.5; P=0.0076), but not p53 (111.1 vs. 36.96; P=0.1536), were overexpressed in the ESCC specimens (see Figure 2A).
Full table
Detection sensitivity and specificity based on the chosen biomarkers
To further evaluate these biomarkers, their detection sensitivity and specificity were calculated. Of the 30 ESCC specimens, 25 showed high expressions according to the optimal cut-off point for EGFR. The calculated sensitivity of EGFR for the identification of ESCC was 83.3% (95% CI, 65.3 to 94.4). Of the 30 non-ESCC specimens, 20 showed low expressions for ESCC. The specificity of EGFR for the identification of ESCC was 70.0% (95% CI, 50.6 to 85.3). The AUC for EGFR was 0.736 (95% CI, 0.61 to 0.84). In relation to p53, the AUC was only 0.61 (95% CI, 0.47 to 0.73), and it accurately identified 14 of 30 ESCC patients (sensitivity, 46.7%; 95% CI, 28.3 to 65.7) and 24 of 30 non-ESCC patients (specificity, 83.3%; 95% CI, 28.3 to 65.7). Twenty-eight of 30 ESCC patients showed high expressions of PCNA. The sensitivity of PCNA for the identification of ESCC was 93.3% (95% CI, 77.9 to 99.2). Of the 30 non-ESCC specimens, 19 showed low expressions for ESCC. The specificity of PCNA for the identification of ESCC was 63.3% (95% CI, 43.9 to 80.1). The AUC for PCNA was 0.80 (95% CI, 0.68 to 0.90). VEGF had an AUC of 0.70 (95% CI, 0.57 to 0.81), and accurately identified 26 of 30 ESCC patients (sensitivity, 86.7%; 95% CI, 69.3 to 96.2) and 18 of 30 non-ESCC patients (specificity, 60.0%; 95% CI, 40.6 to 77.3) (see Figure 2B).
Combination of biomarkers
After the individual analysis of these 4 biomarkers, we found that none of the above-mentioned markers had satisfying AUCs, which illustrated their diagnostic ability. Thus, we investigated whether or not a combination of these biomarkers could improve their diagnostic ability. Due to its low diagnostic ability for ESCC, p53 was excluded from the combinations. As Figure 3A shows, the combination of 2 or 3 markers increased the AUCs. Indeed, the AUC prediction probability increased to 0.852 in the triplicate combination in which the single protein were 0.736 (EGFR), 0.803 (PCNA), and 0.699 (VEGF). There was a significant difference between the predictive ability of EGFR alone, and EGFR combined with PCNA and VEGF Combining VEGF with the other 2 biomarkers elevated the AUC significantly (see Figure 3B). The heatmap shows the IHC expression of each biomarker per patient sample (see Figure 3C).

Expression quantification in the Western blot
As EGFR and PCNA were highly expressed in ESCC tumors in the paraffin-embedded samples, we hypothesized that the levels of EGFR and PCNA in ESCC fresh tissues would be higher than those of adjacent normal tissues. To test this hypothesis, EGFR and PCNA protein expression was analyzed in 5 pairs of excision fresh samples from 5 ESCC patients. The Western blot analysis showed that in 5 pairs of tumor and adjacent normal tissues, EGFR was relatively more highly expressed in 3 tumor tissues than the matched adjacent normal tissues, and PCNA was relatively more highly expressed in 4 tumor tissues than the matched normal tissues (see Figure 4).

Discussion
ESCC is a prevalent malignancy for which no effective therapy exists. Common serum tumor biomarkers, such as CA199, CEA, CA125, and AFP, are not applicable to ESCC (see Table 1). Previous research has shown that PCNA and EGFR are overexpressed in esophageal cancer (32,33). However, no previous study has investigated the diagnostic ability of a single EGFR, p53, PCNA, or VEGF biomarker or a combination of these biomarkers. This is the first study to evaluate the diagnostic ability of these IHC biomarkers for ESCC. We found that combinations of these biomarkers increased the AUCs. Further, the use of a combination of these biomarkers significantly increased their predictive power.
IHC is a reproducible and promising way to assess the expression of oncogenic proteins in cancer tissue samples and to find potential therapeutic targets, and identify patients who are likely to responding well to these specific therapies. Furthermore, IHC is sensitive and specific for mutation of some gene like TP53 (34). Mutation status of these oncogenes predicts response to chemoradiotherapy in esophageal cancer (24). In this study, we described the expression of PCNA, p53, EGFR, and VEGF in 30 ESCC surgical pathology specimens and 30 non-ESCC surgical pathology specimens, and found that PCNA, EGFR, and VEGF were overexpressed, and thus may be important biomarkers of ESCC.
It has been reported that during esophageal neoplastic progression, PCNA appears to increase in intensity (8). In our study, PCNA had the highest sensitivity and AUC of the 4 biomarkers. We also found that while PCNA was fairly sensitive, its specificity was unsatisfactory. Indeed, its specificity was lower than that of EGFR and p53. PCNA appears to be overexpressed in carcinomas; however, it also had high expressions in some active proliferative areas, which may explain why PCNA did not have satisfactory specificity.
According to previous research, EGFR is overexpressed in ESCC. Our findings supported those of previous studies (32). EGFR has a good balance of sensitivity and specificity. Thus, EGFR may be a good candidate as a biomarker of ESCC. Similar to PCNA, VEGF has good sensitivity but low specificity, which weakens its potential in clinical applications.
The p53 protein behaves quite differently to the other 3 biomarkers. There was no statistical significance in the IHC staining scores between the ESCC and non-ESCC groups. Due to its short half-life time, the wild-type p53 protein may barely be detectable by IHC staining. Conversely, aberrant p53 has a longer half-life time, which can be detected by IHC staining (33). In our study, we evaluated the stain strength and range of p53, and found that there was no significant difference between the ESCC and non-ESCC groups. Thus, p53 is not a biomarker of ESCC.
We used computer-assisted imaging to analyze the slides. We used H-scores to reduce the bias for human-based visual semi-quantitative approaches. Computer-assisted IHC analysis is important to achieve robust biomarker scoring to support high-throughput IHC studies. High-throughput genomic, transcriptomic and proteomic studies provide an opportunity to evaluate a large number of novel molecular markers with prognostic significance (35). Computer-assisted imaging minimizes inter-pathologist variations and is more independent and reproducible. In addition, compared to pathologist-based scoring, computer-assisted imaging analysis scoring is a more detailed classification of the stain strength and range, which improves accuracy and comparability. Finally, the results of the Western blot support the hypothesis that the levels of EGFR and PCNA in ESCC fresh tissues were higher than those in adjacent normal tissues, which were consistent with the IHC analysis.
These IHC biomarkers of ESCC in our study are involved in regulating proliferation, cell apoptosis, angiogenesis and metastasis of ESCC cell. Combination of these biomarkers as a panel may be a useful tool for increasing predicted accuracy. So IHC panel might be great helpful for accurate prognostication, development of better surgical care and chemoradiotherapeutic regimes, and identification of novel therapeutics are key areas to address of ESCC (36,37). On the other hand, the IHC panel could be easily adopted by existing pathology service to determine the status for these biomarkers. In our research, we found that the IHC panel containing EGFR, PCNA and VEGF is more sensitivity and specificity than the IHC of the biomarkers separately, which means that the IHC panel might benefit in diagnosis and treatment decisions for ESCC.
Our study was based on 60 clinical samples. The sample size was relatively small; however, it provided high statistical precision, and enabled us to identify the cancer sites most commonly associated with a diagnosis of elevated biomarkers. In this study, only non-ESCC tumors were used as controls for ESCC, as resected normal esophageal tissue is difficult to acquire. However, this might have introduced some bias in relation to common tumor features if the biomarkers are sensitive for both ESCC and non-ESCC. To address this issue, we intend to conduct a large number of validation studies with larger samples sizes and other controls. Another limitation of this study was the lack of complete laboratory data on the extent of biomarker elevations, which may have influenced the association between diagnoses of elevated biomarkers. Due to these limitations and the unexplained elevated biomarkers, we cannot suggest guidelines for the clinical care of patients.
Conclusions
In conclusion, our findings suggest that an initial clinical diagnosis could be based on elevated levels in 3 routinely used proteins (i.e., PCNA, EGFR, and VEGF), as these proteins are biomarkers of ESCC. The incidence risk of many cancers decreases after the use of targeted precision medicine; however, a significantly higher risk remains for ESCC patients. The high detection rate of PCNA, EGFR, and VEGF, alone or in combination, indicate that they are independent or associated with each other in the differentiation and proliferation of ESCC. These biomarkers represent potentially promising targets for molecular therapy and molecular imaging for ESCC.
Acknowledgments
The authors would like to thank Mr. Yucheng Cao for his helpful technical support and Mrs. Yuhong Ji for providing the standard for the high-resolution IHC scanner.
Funding: This research was funded by the National Key R&D Program of China (2018YFC091060), the National Natural Science Foundation of China (81871382) and Key Realm R&D Program of Guangdong Province (2018B030337001).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-2950
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-2950
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-2950). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional research ethics board of the Fifth Affiliated Hospital, Sun Yat-sen University (No. 2019 K20-1). Because of the retrospective nature of the study, patient consent for inclusion was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Shang QX, Yang YS, Hu WP, et al. Clinical and prognostic significance of preoperative lymphocyte-monocyte ratio, neutrophil-lymphocyte ratio and neutrophil-monocyte ratio on esophageal squamous cell carcinoma patients. Transl Cancer Res 2020;9:3903-14. [Crossref]
- Lagergren J, Smyth E, Cunningham D, et al. Oesophageal cancer. Lancet 2017;390:2383-96. [Crossref] [PubMed]
- Wang SC. PCNA: a silent housekeeper or a potential therapeutic target? Trends Pharmacol Sci 2014;35:178-86. [Crossref] [PubMed]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell 2007;129:665-79. [Crossref] [PubMed]
- Kimos MC, Wang S, Borkowski A, et al. Esophagin and proliferating cell nuclear antigen (PCNA) are biomarkers of human esophageal neoplastic progression. Int J Cancer 2004;111:415-7. [Crossref] [PubMed]
- Tan Z, Wortman M, Dillehay KL, et al. Small-molecule targeting of proliferating cell nuclear antigen chromatin association inhibits tumor cell growth. Mol Pharmacol 2012;81:811-9. [Crossref] [PubMed]
- Muller R, Misund K, Holien T, et al. Targeting proliferating cell nuclear antigen and its protein interactions induces apoptosis in multiple myeloma cells. PLoS One 2013;8:e70430 [Crossref] [PubMed]
- Yu YL, Chou RH, Liang JH, et al. Targeting the EGFR/PCNA signaling suppresses tumor growth of triple-negative breast cancer cells with cell-penetrating PCNA peptides. PLoS One 2013;8:e61362 [Crossref] [PubMed]
- Surget S, Khoury MP, Bourdon JC. Uncovering the role of p53 splice variants in human malignancy: a clinical perspective. Onco Targets Ther 2013;7:57-68. [PubMed]
- Gao YB, Chen ZL, Li JG, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet 2014;46:1097-102. [Crossref] [PubMed]
- Lin DC, Hao JJ, Nagata Y, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet 2014;46:467-73. [Crossref] [PubMed]
- Su Y, Yin L, Liu R, et al. Promoter methylation status of MGMT, hMSH2, and hMLH1 and its relationship to corresponding protein expression and TP53 mutations in human esophageal squamous cell carcinoma. Med Oncol 2014;31:784. [Crossref] [PubMed]
- Zhang SS, Huang QY, Yang H, et al. Correlation of p53 status with the response to chemotherapy-based treatment in esophageal cancer: a meta-analysis. Ann Surg Oncol 2013;20:2419-27. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Rehmani HS, Issaeva N. EGFR in head and neck squamous cell carcinoma: exploring possibilities of novel drug combinations. Ann Transl Med 2020;8:813. [Crossref] [PubMed]
- Walker F, Abramowitz L, Benabderrahmane D, et al. Growth factor receptor expression in anal squamous lesions: modifications associated with oncogenic human papillomavirus and human immunodeficiency virus. Hum Pathol 2009;40:1517-27. [Crossref] [PubMed]
- Stuttfeld E, Ballmer-Hofer K. Structure and function of VEGF receptors. IUBMB Life 2009;61:915-22. [Crossref] [PubMed]
- Holmes K, Roberts OL, Thomas AM, et al. Vascular endothelial growth factor receptor-2: structure, function, intracellular signalling and therapeutic inhibition. Cell Signal 2007;19:2003-12. [Crossref] [PubMed]
- Yan T, Cui H, Zhou Y, et al. Multi-region sequencing unveils novel actionable targets and spatial heterogeneity in esophageal squamous cell carcinoma. Nat Commun 2019;10:1670. [Crossref] [PubMed]
- Chu LY, Peng YH, Weng XF, et al. Blood-based biomarkers for early detection of esophageal squamous cell carcinoma. World J Gastroenterol 2020;26:1708-25. [Crossref] [PubMed]
- Makino T, Yamasaki M, Miyata H, et al. p53 Mutation status predicts pathological response to chemoradiotherapy in locally advanced esophageal cancer. Ann Surg Oncol 2010;17:804-11. [Crossref] [PubMed]
- Tomasini P, Greillier L. Targeted next-generation sequencing to assess tumor mutation burden: ready for prime-time in non-small cell lung cancer? Transl Lung Cancer Res 2019;8:S323-6. [Crossref] [PubMed]
- Jin H, Xu M, Padakanti PK, et al. Preclinical evaluation of the novel monoclonal antibody H6-11 for prostate cancer imaging. Mol Pharm 2013;10:3655-64. [Crossref] [PubMed]
- Jin H, Hallstrand TS, Daly DS, et al. A halotyrosine antibody that detects increased protein modifications in asthma patients. J Immunol Methods 2014;403:17-25. [Crossref] [PubMed]
- Allred DC, Bustamante MA, Daniel CO, et al. Immunocytochemical Analysis of Estrogen Receptors in Human Breast Carcinomas: Evaluation of 130 Cases and Review of the Literature Regarding Concordance With Biochemical Assay and Clinical Relevance. Arch Surg 1990;125:107-13. [Crossref] [PubMed]
- Ishibashi H, Suzuki T, Suzuki S, et al. Sex steroid hormone receptors in human thymoma. J Clin Endocrinol Metab 2003;88:2309-17. [Crossref] [PubMed]
- Hanley JAM. B J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982;143:29-36. [Crossref] [PubMed]
- Perkins NJ, Schisterman EF. The Inconsistency of “Optimal” Cutpoints Obtained using Two Criteria based on the Receiver Operating Characteristic Curve. Am J Epidemiol 2006;163:670-5. [Crossref] [PubMed]
- Gibault L, Metges JP, Conan-Charlet V, et al. Diffuse EGFR staining is associated with reduced overall survival in locally advanced oesophageal squamous cell cancer. Br J Cancer 2005;93:107-15. [Crossref] [PubMed]
- Purdie CA, O'Grady J, Piris J, et al. p53 expression in colorectal tumors. Am J Pathol 1991;138:807-13. [PubMed]
- Guedes LB, Almutairi F, Haffner MC, et al. Analytic, Preanalytic, and Clinical Validation of p53 IHC for Detection of TP53 Missense Mutation in Prostate Cancer. Clin Cancer Res 2017;23:4693-703. [Crossref] [PubMed]
- Suresh A, Vannan M, Kumaran D, et al. Resistance/response molecular signature for oral tongue squamous cell carcinoma. Dis Markers 2012;32:51-64. [Crossref] [PubMed]
- Ong CA, Shapiro J, Nason KS, et al. Three-gene immunohistochemical panel adds to clinical staging algorithms to predict prognosis for patients with esophageal adenocarcinoma. J Clin Oncol 2013;31:1576-82. [Crossref] [PubMed]
- Yu Y, Li Z, Huang C, et al. Integrated analysis of genomic and transcriptomic profiles identified a prognostic immunohistochemistry panel for esophageal squamous cell cancer. Cancer Med 2020;9:575-85. [Crossref] [PubMed]
(English Language Editors: L. Huleatt)


