
High expression of PYCARD is an independent predictor of unfavorable prognosis and chemotherapy resistance in glioma
Introduction
Glioma is among the most common human central nervous malignancy; the clinical prognosis is extremely poor due to acquired resistance to temozolomide (TMZ) chemotherapy, and is usually partitioned into low-grade glioma (LGG) and high-grade glioma (HGG) by pathological grades (1,2). Unfortunately, most LGG cases inevitably progress to HGG. Glioblastomas/glioblastoma multiforme (GBMs) are mostly HGG, and patients with GBM demonstrate a short survival time and poor prognosis(3). Currently, surgery and chemotherapy are the primary therapeutic methods for glioma treatment. Gliomas demonstrate multiple modes of resistance and temozolomide (TMZ) resistance one of the most influential contributing factors to limit the efficacy of chemotherapy in glioma patients (4-7). Furthermore, because chemoresistance of glioma cells may significantly influence the curative effect of glioma patients, a more extensive clarification of the etiology inherent in glioma development and chemosensitivity is critical.
The TMZ resistance of glioma is a complex process. Multiple studies have shown that TMZ chemoresistance is related to various biological processes, including apoptosis, proliferation, migration, and invasion (8). Several studies have revealed that O6-methylguanine-DNA methyltransferase (MGMT) and DNA repair protein are related to drug resistance (9-12). Similarly, TMZ regulates cellular processes, such as proliferation, apoptosis, migration, and invasion through various signaling pathways, including EGFR/AKT/PTEN signaling pathway, protein kinase C signaling pathway, P53 signaling pathway, retinoblastoma signaling pathway, vascular endothelial growth factors, integrin signaling pathway, and protein transporter (13). However, the underlying molecular mechanism of TMZ chemoresistance is unclear. Molecular mechanisms of TMZ chemoresistance are essential to improve therapeutic strategies for malignant gliomas.
In this study, we used bioinformatics analysis to examined resistance cells (HS683R, LNZ308R, U87R, and LN229R) and compared them with the parental cells (HS683, LNZ308, U87, and LN229): we found that PYD and CARD domain containing (PYCARD) was upregulated only in resistant cells (HS683R, LNZ308R, and LN229R). PYCARD is a protein-coding gene that mediates the assembly of large signaling complexes in the inflammatory and apoptotic signaling pathways via the activation of caspase (14,15). The mRNA and protein expression levels of ASC/TMS1 (PYCARD) in gastric cancer tissues were lower than those in normal gastric mucosal tissues and associated with poor prognosis of patients with gastric cancer(16). Remarkably, we found that PYCARD was upregulated in TMZ-resistant cell lines and glioma tissue, correlated with poor prognosis, and promoted glioma cell proliferation and migration. However, the specific mechanism of PYCARD remains elusive.
Over the past decade, research has demonstrated that immune factors are associated with increased glioma risk and immune defects (17). Treatment with immune checkpoint inhibitors, including programmed death-ligand (PD-L1), has improved glioma patient outcomes (18,19). In the glioma microenvironment, infiltrating immune cells have been demonstrated to benefit tumor progression (20). For example, cluster of differentiation (CD)30 ligand deficiency accelerates glioma progression by promoting the formation of tumor immune microenvironment (21). However, the association between the expression of PYCARD and the immune microenvironment has yet to be determined.
Long non-coding RNA (lncRNA) is a non-coding RNA longer than 200 nucleotides and has no protein-coding function. Abundant studies have indicated that lncRNAs carry out many biological functions in glioma, including cell proliferation, invasion, migration, TMZ chemoresistance, metastasis, and apoptosis, by altering the expression of target genes (22-24). However, no reports concerning the relationships between PYCARD and lncRNAs gene expression have yet been published.
Herein, we clarify the specific mechanism of PYCARD in TMZ resistance and the association between the expression of PYCARD, immune microenvironment, and lncRNAs. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/atm-21-2346).
Methods
Clinical specimen collection
All 114 human glioma tissue samples and patient clinical information were obtained from the Department of Neurosurgery at Jiangxi Provincial People’s Hospital Affiliated to Nanchang University, China. Fifty normal brain tissues samples and glioma tissues specimens, representing 62 grade IV GBM, 30 grade III, and 22 grade II cases, were classified by 2 neurosurgeons according to the 2010 World Health Organization (WHO) criteria for glioma. Samples were frozen in liquid nitrogen immediately after surgical resection of the patient’s tumor. The enrolled patients were followed up by telephone. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The present study was approved by the Ethics Committee of the Jiangxi Provincial People’s Hospital Affiliated to Nanchang University and informed consent was taken from all the patients.
Bioinformatic analysis
The expression profiles obtained from the Gene Expression Omnibus (GEO) data set (GSE53014 and GSE113510) (https://www.ncbi.nlm.nih.gov/geo/) were analyzed with the “limma” and “RobustRankAggreg” package in R (version 4.0.2, R Foundation for Statistical Computing). Next, we performed survival analyses based on the expression of PYCARD in The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/). We used the Chinese Glioma Genome Atlas (CGGA) (http://www.cgga.org.cn/) data base to further validate the expression of PYCARD and the survival rate in glioma.
PYCARD-related lncRNA was found by using Spearman correlation analysis (R >0.3 and P value <0.05). Univariate Cox and Kaplan-Meier analyses were used to identify those lncRNAs with clinical prognostic value. Then, Multivariate analysis was conducted by cox-regression analysis these lncRNAs to build a risk prediction model. The prognostic gene signature was determined according to the following formula: risk score = (Coefficient lncRNA1 × expression of lncRNA1) + (Coefficient lncRNA2 × expression of lncRNA2) + … + (Coefficient lncRNAn × expression lncRNA). The “survival” package was used to determine the optimal cutoff value and assign patients to high and low risk groups based on risk scores.
Correlation was analyzed to explore the relation of PYCARD genes with immune cell infiltration by using “ggplot2”, “ggpubr”, “ggExtra”, “limma”, “estimate”, “preprocessCore”, and “CIBERSORT” packages.
The correlation between of PYCARD genes and tumor mutation burden (TMB) was analyzed using the TCGA TMB database, while the correlation between PYCARD genes and microsatellite instability (MSI) was analyzed by using the “fmsb” package.
Gene set enrichment analysis (GSEA)
GSEA was performed though the GSEA software (GSEA version 4.0.2). Gene sets with a standard P value <0.05 were considered to be significantly enriched.
Cell culture
We acquired Human U-251MG (U251) and LN-229 (LN229) glioma cell lines from the Cell Repository, Chinese Academy of Sciences (Shanghai, China). Glioma cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Corning, VA, USA) complemented with 10% fetal bovine serum (FBS; Corning, VA, USA). TMZ-resistant cell lines, U251R and LN229R, were generated through chronic TMZ exposure (48 hours every 2 weeks) as previously described (25-27). To maintain the LN229R and U251R resistance phenotype, cells were regularly exposed to TMZ (50 µg/mL; Sigma-Aldrich, San Francisco, CA, USA) during routine culturing. All cells were cultured in a humidified incubator with 5% carbon dioxide (CO2) and 95% air at 37 °C.
Cell lentiviral transduction
Stable cell lines were generated by lentiviral transduction. The plasmid construction and lentivirus package were performed commercially (Genechem Co. Ltd., Shanghai, China). We plated glioma cells in a 6-well plate with a density greater than 40–50%. Cell transfection was performed according to the GeneChem (Shanghai, China) manufacturers’ instruction. The target sequences of short hairpin RNAs (shRNAs) are listed in Table S1. Gene transfection efficiency was evaluated by quantitative real-time polymerase chain (qRT-PCR) or western blotting.
Quantitative real-time polymerase PCR
From the glioma tissue samples and cells, total RNA was extracted with TRIzol according to the manufacturer’s instructions (Takara Bio, Shiga, Japan). First-strand complement DNA (cDNA) for genes was generated by using the First Strand cDNA Synthesis Kit (Takara Bio, Shiga, Japan). The determination of messenger RNA (mRNA) levels was determined by using SYBR Green Master Mix (Takara Bio, Shiga, Japan) and performed on a 7500 Fast Real-time PCR System (Applied Biosystem, Foster City, CA, USA). GAPDH was used as the endogenous control. The quantitative PCR primers are listed in Table S2. The relative expression was calculated through relative quantification (2−∆∆Ct), and the data were presented as the mean ± SD of at least 3 independent experiments.
Western blotting and antibodies
Western blotting assay was executed according to according to standard protocols. Antibody supply information was as follows: PYCARD, Vimentin, N-cadherin, E-cadherin, Ki67, and Actin (Cell Signaling Technology, MA, USA). Subsequently, Membranes were washed for 0.5 h and incubated for 2 h with the appropriate secondary antibodies (Cell Signaling Technology, MA, USA). Membranes were then detected and visualized by chemiluminescence. The antibody information is listed in Table S3.
Cell viability and cell migration assays
Cell viability was evaluated by MTT assay. Glioma cells were treated with 50 µg/ml TMZ for 1–5 days (26). Glioma cell growth was monitored by measuring the absorbance at 450 after 1-h incubation in 10% cell counting kit-8 (CCK-8) (Beyotime, China) solution according to the manufacturer’s instructions.
In the migration assays, cells were incubated in 6-well plates with an 8-mm pore size polycarbonate membrane for 48 h. The cells at the bottom of the chamber were wiped with a cotton swab, and the cells on the surface of the lower membrane were fixed with methanol for 0.5 h, and then stained with 0.1% crystal violet.
Immunofluorescence staining
Glioma cells were seeded overnight on a glass cover glass (0.17 mm thick and 14 mm diameter) in a 6-well plate. The cells were washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 0.5 min, permeated with 0.1%Triton X-100 for 10 min, and sealed with 2% bovine serum albumin for 1 h. The cells were then cultured overnight with PYCARD antibody at 4 °C. After 3 rinses with PBS, appropriate fluorescent secondary antibodies were added to the cell samples and cultured at room temperature for 2 h. The nucleus was counterstained by DAPI. Finally, sections were detected under a Carl Zeiss LSM 700 confocal microscope (Zeiss, Jena, Germany).
Statistical analysis
All bioinformatics and statistical analyses were performed using R statistical software. For cell quantification, an unpaired Student’s t-test was used in GraphPad Prism 8 software. The standard error of the mean (SEM) was applied to calculate the error bars. P<0.05 is considered as statistically significant.
Results
PYCARD was highly expressed in TMZ-resistant cell lines and glioma tissue and correlated with poor prognosis
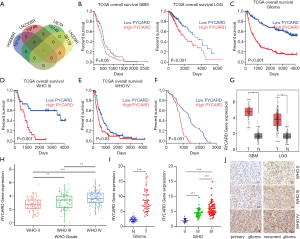
The potential involvement of the gene in the regulation of TMZ chemoresistance in glioma was determined. Next, we analyzed resistance cells (HS683R, LNZ308R, U87R, and LN229R) and compared them with parental cells (HS683, LNZ308, U87, and LN229) using data derived from the GEO database (GSE53014 and GSE113510; https://www.ncbi.nlm.nih.gov/geo/). Using bioinformatics analysis, we found that PYCARD was upregulated only in resistance cells (HS683R, LNZ308R, and LN229R; Figure 1A). Next, we further attempted to study the function of PYCARD expression in glioma patient survival. Survival analysis from TCGA database showed that an elevated level of PYCARD was associated with unfavorable prognosis in GBM and LGG (Figure 1B). A similar result was observed in all the WHO grade (primary glioma) and grades III–IV glioma based on CGGA database analysis (Figure 1C-E). We further analyzed the expression of PYCARD in correlation with patient survival times based on the first surgical procedure performed in our center for each patient in our clinical cohorts using the Kaplan-Meier method (Figure 1F). Further analysis of TCGA data showed that compared with normal brain tissue, PYCARD was highly expressed in GBM and LGG (Figure 1G). Likewise, CGGA data revealed that the mRNA level of PYCARD was significantly increased in HGG compared to LGG (Figure 1H). To confirm the increased expression of PYCARD in glioma tissues, we measured the expression of PYCARD using qRT-PCR in clinical glioma specimens (Figure 1I). Similar results were also obtained from immunohistochemical staining. PYCARD was highly expressed in HGG compared to LGG, and PYCARD expression was more highly expressed in recurrent gliomas than in primary gliomas (Figure 1J). Overall, these results indicated PYCARD to be upregulated in TMZ-resistant cell lines and glioma tissue and to be correlated with poor prognosis.
PYCARD promoted glioma cell proliferation and migration
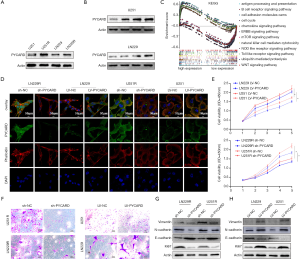
As PYCARD was overexpressed in glioma and related to poor prognosis, we next conducted functional experiments to investigate the potential biological function of PYCARD. First, western blotting was applied to verify the high expression of PYCARD in resistance cells (U251R and LN229R) compared with the parental cells (U251 and LN229) (Figure 2A). PYCARD protein expression levels were significantly elevated with increased concentration of TMZ (0, 25, 50, and 100 µg/mL) in both parental cell lines (Figure 2B). GSEA results showed that proliferation, metastasis, and immunity were closely correlated with high expression levels of PYCARD (Figure 2C). U251R or LN229R cells were transfected with PYCARD shRNA lentiviral vectors (sh-PYCARD) or Control shRNA lentiviral vectors (sh-NC), lentiviral vectors to knockdown PYCARD expression, and U251 or LN229 cells were transfected with Control lentiviral activation vectors (LV-NC) or PYCARD lentiviral activation vectors (LV-PYCARD), lentiviral vectors to generate stable PYCARD-overexpressed cells to assess the role of PYCARD in glioma cell proliferation and migration, respectively. Transfection efficiency was measured by immunofluorescence (Figure 2D). CCK-8 results showed that PYCARD overexpression significantly increased the activity of cells after TMZ treatment (50 µg/mL). Conversely, the loss of PYCARD decreased cell viability (Figure 2E). Cell migration assays results showed that overexpression of PYCARD significantly elevated cell migration ability after TMZ treatment, while the loss of PYCARD decreased cell migration ability (Figure 2F). We then examined proliferation-related proteins and metastasis-related proteins using western blotting (Figure 2G,H). Overall, these data demonstrate that PYCARD promotes glioma cell proliferation and migration.
Association of PYCARD expression with the tumor microenvironment
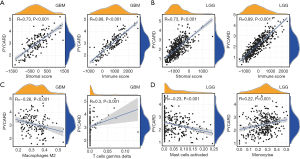
We determined whether PYCARD expression was correlated with tumor microenvironment (TME) in GBM and LGG by calculating the coefficient of PYCARD expression and TME. The outcome revealed that PYCARD expression had a significant correlation with stromal and immune cells in GBM and LGG (Figure 3A,B). Furthermore, PYCARD expression showed a significantly positive correlation with gamma delta T cells but a negative correlation with M2 macrophages in GBM (Figure 3C). Similarly, PYCARD expression demonstrated a significantly positive association with the monocytes but a negative correlation with activated mast cells in LGG (Figure 3D). These findings strongly indicated that PYCARD affects patient survival through interacting with TME in GBM and LGG.
PYCARD-related lncRNA signature for glioma
Although PYCARD is involved in glioma cell proliferation and migration, the exact regulatory mechanism has yet to be elucidated. Thus, the relationship between PYCARD and lncRNAs from TCGA dataset was analyzed. First, the construction of a coexpression network for PYCARD-lncRNAs was extracted from TCGA dataset (R>0.3; P<0.001). Univariate Cox regression analysis was applied using 14,142 PYCARD-associated lncRNAs in GBM (P<0.05) and 14,142 PYCARD-associated lncRNAs in LGG (P<0.001; Table S4 and Table S5). After initial univariate Cox regression analysis to screen prognostic lncRNAs, multivariate Cox regression analysis was applied based on 8 PYCARD-associated lncRNAs in GBM and 11 PYCARD-associated lncRNAs in LGG (Table S6 and Table S7). Three lncRNAs in GBM and four lncRNAs in LGG had a predictive value for glioma patients (Figure 4A). To demonstrate the relationship between lncRNAs and PYCARD, we determined the expression level of lncRNAs and PYCARD using qRT-PCR in 114 glioma tissues. The Pearson coefficient was adopted to estimate the correlation between lncRNAs and PYCARD. We found that AC122129.1 and AC017104.1 gene expression was negatively correlated with PYCARD genes, while AL357992.1 gene expression was positively associated with PYCARD genes in GBM (Figure 4B,C,D). Further, we used a risk score method to develop a PYCARD-associated lncRNA signature. We divided the GBM patients into a low-risk group and a high-risk group, and differences in risk score based on AC122129.1, AC017104.1, and AL357992.1 expression were evaluated. This revealed that the high-risk GBM patients had a shorter median overall survival than did the low-risk GBM patients (Figure 4E). Subsequently, AC122129.1, AC017104.1, and AL357992.1 were considered to be independent prognostic factors for GBM patients: AC017104.1 and AL357992.1 were unfavorable factors, and AC122129.1 was a favorable factor (Figure 4F,G,H).
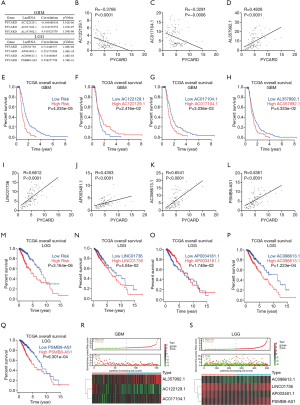
Similarly, we found that LINC01736, AP003481.1, AC098613.1, and PSMB8-AS1 gene expression positively correlated with PYCARD genes in 114 glioma tissues (Figure 4I-L). We also assessed the differences in the risk score using LINC01736, AP003481.1, AC098613.1, and PSMB8-AS1 expression. The high-risk LGG patients had a shorter median overall survival than did the low-risk LGG patients (Figure 4M). Subsequently, LINC01736, AP003481.1, AC098613.1, and PSMB8-AS1 were found to be independent prognostic factors for LGG patients: LINC01736, AC098613.1, and PSMB8-AS1 were unfavorable factors, whereas AP003481.1 was a favorable factor (Figure 4M,N,O,P,Q).
Further, these results were validated in TCGA datasets, with GBM and LGG patients being enrolled for the validation of the lncRNA signature (Figure 4R,S). These findings strongly suggest that lncRNAs may be an independent prognostic indicator for GBM and LGG patients.
GSEA analysis
Among the gene sets, several signaling pathways were highly enriched in GBM, including the EGF receptor family (ERBB) signaling pathway, glioma, inositol phosphate metabolism, and phosphatidylinositol signaling system (Figure 5A,B,C,D).
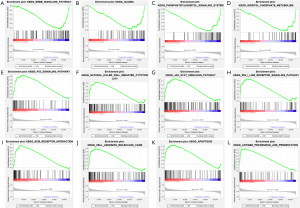
Likewise, GSEA indicated a significant enrichment of hallmark cancer-related pathways in the high-risk group based on the LGG database. Among the gene sets, several signaling pathways were well established in LGG, including P53 signaling pathway, natural killer cell-mediated, JAK/STAT signaling pathway, RIG I-like receptor signaling pathway, extracellular matrix (ECM) receptor interaction, cell adhesion molecules cams, apoptosis, and antigen processing and presentation (Figure 5E,F,G,H,I,J,K,L). Our data indicated that lncRNAs might play an essential role in the modulation of glioma progression.
Pan-cancer analysis of PYCARD mRNA expression and prognosis
The expression level of PYCARD mRNA in pan-carcinoma was analyzed in TCGA. The results indicated that compared with their respective normal groups, PYCARD expression was higher in cancer groups, including prostate adenocarcinoma (PRAD), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma (CESC), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), cholangiocarcinoma (CHOL), esophageal carcinoma (ESCA), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), and uterine corpus endometrial carcinoma (UCEC) (Figure 6A). The expression of PYCARD in different cancer types in was compared using Kaplan-Meier survival curves. Similarly, we also found that PYCARD was an adverse prognostic factor for KIRC and Acute Myeloid Leukemia (LAML) (Figure 6B,C). PYCARD expression associated positively with worsening tumor grade in KIRC (Figure 6D).
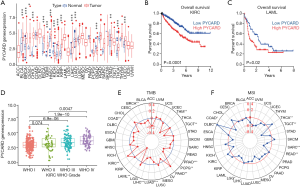
TMB data downloaded from TCGA database were used to evaluate PYCARD mRNA expression levels. Using recently published MANTIS (Microsatellite Analysis for Normal-Tumor InStability) software, we analyzed whole-exome data from TCGA sources across 33 cancers. Within a subset of these cancers, we assessed the association of MSI with PYCARD expression. According to the radar plot, high TMB inhibited the expression of PYCARD in thymoma (THYM), testicular germ cell tumors (TGCT), Prostate adenocarcinoma (PRAD), pheochromocytoma and paraganglioma (PCPG), lung squamous cell carcinoma (LUSC), LIHC, LAML, colon adenocarcinoma (COAD), and BRCA as compared to (pancreatic adenocarcinoma) PAAD and KIRC. However, the regulatory relationships in other cancers were not statistically significant (Figure 6E). MSI may also enhance PYCARD. The expression of PYCARD was promoted in THCA and lymphoid neoplasm diffuse large B-cell lymphoma (DLBC) but inhibited in testicular germ cell tumors (TGCT), sarcoma (SARC), rectum adenocarcinoma (READ), LGG, COAD (Figure 6F).
Discussion
The TMZ resistance of glioma is a complex process, and multiple studies have shown that TMZ chemoresistance is related to various biological processes (2,12,13). However, the underlying molecular mechanism of TMZ chemoresistance remains unclear. Molecular mechanisms of TMZ chemoresistance are essential to improving the therapeutic strategies for malignant gliomas.
In the present study, we first analyzed resistance cells (HS683R, LNZ308R, U87R, and LN229R) and compared them with parental cells (HS683, LNZ308, U87, and LN229). Using bioinformatics analysis (GSE53014 and GSE113510), we found that PYCARD was upregulated only in resistance cells (HS683R, LNZ308R, and LN229R), but was downregulated in resistance cell U87R. PYCARD is a protein-coding gene that mediates the assembly of large signaling complexes in the inflammatory and apoptotic signaling pathways via the activation of caspase (14,15). This proapoptotic activity is mediated predominantly through the activation of caspase-9 (28,29). Methylation of PYCARD (ASC/TMS1) promoter is associated with poor prognosis of patients with gastric cancer (16). Surprisingly, proapoptotic PYCARD genes are highly expressed in the resistance cells. However, how and why this occurs is crucial information that has yet to be clarified. Therefore, we performed a functional study of PYCARD.
Recent studies have found PYCARD deficiency suppresses proliferation and prevents medulloblastoma incidence (30). We also found that overexpression of PYCARD significantly promoted cell proliferation and migration ability after TMZ treatment; conversely, the loss of PYCARD decreased cell proliferation and migration ability. However, no noticeable apoptotic and half-maximal inhibitory concentration (IC50) changes were observed in our research (30-33). This is inconsistent with the previous results (32), and this discrepancy is due to cancer heterogeneity and the complex network regulating mRNA. Nonetheless, this deserves further exploration.
Survival analysis from TCGA database indicated that the higher expression of PYCARD was correlated with poor prognosis in GBM and LGG. A similar result was observed in all WHO grade (primary glioma) and WHO III–IV glioma based on CGGA database analysis. Our survival analyses revealed that PYCARD expression was significantly associated with poor clinical outcomes based on patients’ surgery at Jiangxi Provincial People’s Hospital Affiliated to Nanchang University. However, PYCARD had no predictive significance for WHO grade II glioma (n=8), which may contribute to a limited number of glioma cases.
In recent years, studies have found that immune cells in the TME can influence the survival of patients with glioma (17,20,34). Immune checkpoint proteins PD-L1, CTLA-4, TIGIT and TIM-3 are significantly upregulated with the hypomethylation of promoters. The increased levels of immune checkpoint molecules may be one of the causes of repressed activation and function of immune cells in the tumor microenvironment. Therefore, investigating the association between TME and PYCARD expression could be extremely valuable. Our findings that PYCARD expression had significant correlations with stromal and immune cells in GBM and LGG. PYCARD expression was also significantly correlated with gamma delta T cells but negatively correlated M2 macrophages in GBM. Similarly, PYCARD expression was significantly associated with monocytes but negatively correlated with activated mast cells in LGG. Thus, immunity plays an important role in the treatment resistance of TMZ chemotherapy.
Although we found that PYCARD was involved in glioma cell proliferation and migration, the actual regulatory mechanism remains unknown. LncRNA is known to figure prominently in gene expression regulation, with a single target gene being influenced by one or more lncRNAs (23,24). Thus, we attempted to investigate the complicated relationship between PYCARD and lncRNAs. Three PYCARD-associated lncRNAs in GBM and 4 PYCARD-associated lncRNAs in LGG were found to have predictive value for glioma patients. Numerous PYCARD-associated lncRNAs were identified in LGG, perhaps due to our stringent screening criteria (P<0.001). LINC01736, AP003481.1, AC098613.1, and PSMB8-AS1 were considered to be independent prognostic factors for LGG patients, whereas AC122129.1, AC017104.1, and AL357992.1 were considered to be independent prognostic factors for GBM patients. GSEA indicated that lncRNAs might play an essential role in the modulation of glioma progression. However, future studies should be conducted to verify this finding.
The expression level of PYCARD mRNA was analyzed in TCGA to determine the expression of PYCARD in various cancers. We found that PYCARD expression was higher in most of the cancer types. Perhaps due to the heterogeneity of the cancer types, PYCARD was only found to be a prognostic factor in KIRC and LAMA.
From the aspect of its mechanism of action, the following findings are worth pointing out. DNA methylation is a biological process always represses the expression of tumor-suppressive genes in many types of cancer (35). Previous studies have shown that PYCARD is inhibitor by DNA methylation in human breast tumors (36). Aberrant methylation and down-regulation of PYCARD by DNA methyltransferase 1 (Dnmt1), Dnmt3b and Dnmt1/Dnmt3a in human glioblastoma (30,37-40). However, the detailed molecular mechanism behinds the resistance remains unclear, and further research to explore this area is warranted.
Overall, our study provides evidence of the diagnostic and prognostic value of PYCARD expression in glioma patients. The expression of PYCARD plays a crucial role in the resistance to chemotherapy and immunotherapy.
Conclusions
The increased expression level of PYCARD might be a potential biomarker for the diagnosis and prognosis of glioma. Moreover, PYCARD might participate in the TMZ resistance of glioma via the proliferation and migration of the corresponding signaling pathways. Additionally, the expression level of PYCARD has close association with lncRNAs and TME. LncRNAs may be an independent prognostic indicator for glioma patients.
Acknowledgments
We are very thankful for all the participants of the present study. We also thank the reviewers for their valuable advice.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-2346
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-2346
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-2346). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The present study was approved by the Ethics Committee of the Jiangxi Provincial People’s Hospital Affiliated to Nanchang University and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- van den Bent MJ, Weller M, Wen PY, et al. A clinical perspective on the 2016 WHO brain tumor classification and routine molecular diagnostics. Neuro Oncol 2017;19:614-24. [Crossref] [PubMed]
- Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol 2006;1:97-117. [Crossref] [PubMed]
- Pearson JRD, Regad T. Targeting cellular pathways in glioblastoma multiforme. Signal Transduct Target Ther 2017;2:17040. [Crossref] [PubMed]
- Saito T, Sugiyama K, Takeshima Y, et al. Prognostic implications of the subcellular localization of survivin in glioblastomas treated with radiotherapy plus concomitant and adjuvant temozolomide. J Neurosurg 2018;128:679-84. [Crossref] [PubMed]
- Julka PK, Sharma DN, Mallick S, et al. Postoperative treatment of glioblastoma multiforme with radiation therapy plus concomitant and adjuvant temozolomide: A mono-institutional experience of 215 patients. J Cancer Res Ther 2013;9:381-6. [Crossref] [PubMed]
- Pollack IF, Jakacki RI, Blaney SM, et al. Phase I trial of imatinib in children with newly diagnosed brainstem and recurrent malignant gliomas: a Pediatric Brain Tumor Consortium report. Neuro Oncol 2007;9:145-60. [Crossref] [PubMed]
- Prados MD, Chang SM, Butowski N, et al. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol 2009;27:579-84. [Crossref] [PubMed]
- Hombach-Klonisch S, Mehrpour M, Shojaei S, et al. Glioblastoma and chemoresistance to alkylating agents: Involvement of apoptosis, autophagy, and unfolded protein response. Pharmacol Ther 2018;184:13-41. [Crossref] [PubMed]
- Fan CH, Liu WL, Cao H, et al. O6-methylguanine DNA methyltransferase as a promising target for the treatment of temozolomide-resistant gliomas. Cell Death Dis 2013;4:e876 [Crossref] [PubMed]
- Oldrini B, Vaquero-Siguero N, Mu Q, et al. MGMT genomic rearrangements contribute to chemotherapy resistance in gliomas. Nat Commun 2020;11:3883. [Crossref] [PubMed]
- Yi GZ, Huang G, Guo M, et al. Acquired temozolomide resistance in MGMT-deficient glioblastoma cells is associated with regulation of DNA repair by DHC2. Brain 2019;142:2352-66. [Crossref] [PubMed]
- Erasimus H, Gobin M, Niclou S, et al. DNA repair mechanisms and their clinical impact in glioblastoma. Mutat Res Rev Mutat Res 2016;769:19-35. [Crossref] [PubMed]
- Balca-Silva J, Matias D, Carmo AD, et al. Cellular and molecular mechanisms of glioblastoma malignancy: Implications in resistance and therapeutic strategies. Semin Cancer Biol 2019;58:130-41. [Crossref] [PubMed]
- Pierini R, Juruj C, Perret M, et al. AIM2/ASC triggers caspase-8-dependent apoptosis in Francisella-infected caspase-1-deficient macrophages. Cell Death Differ 2012;19:1709-21. [Crossref] [PubMed]
- Franklin BS, Latz E, Schmidt FI. The intra- and extracellular functions of ASC specks. Immunol Rev 2018;281:74-87. [Crossref] [PubMed]
- Wu L, Zhang C, Wang X, et al. Methylation of ASC/TMS1 promoter is associated with poor prognosis of patients with gastric cancer. Clin Transl Oncol 2016;18:296-303. [Crossref] [PubMed]
- Zhong QY, Fan EX, Feng GY, et al. A gene expression-based study on immune cell subtypes and glioma prognosis. BMC Cancer 2019;19:1116. [Crossref] [PubMed]
- Khasraw M, Reardon DA, Weller M, et al. PD-1 Inhibitors: Do they have a Future in the Treatment of Glioblastoma? Clin Cancer Res 2020;26:5287-96. [Crossref] [PubMed]
- Guo H, Wang R, Wang D, et al. Deliver anti-PD-L1 into brain by p-hydroxybenzoic acid to enhance immunotherapeutic effect for glioblastoma. J Control Release 2020;320:63-72. [Crossref] [PubMed]
- Huang S, Song Z, Zhang T, et al. Identification of Immune Cell Infiltration and Immune-Related Genes in the Tumor Microenvironment of Glioblastomas. Front Immunol 2020;11:585034 [Crossref] [PubMed]
- Duan J, Gao Y, Zhang X, et al. CD30 ligand deficiency accelerates glioma progression by promoting the formation of tumor immune microenvironment. Int Immunopharmacol 2019;71:350-60. [Crossref] [PubMed]
- Nair L, Chung H, Basu U. Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nat Rev Mol Cell Biol 2020;21:123-36. [Crossref] [PubMed]
- Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol 2019;21:542-51. [Crossref] [PubMed]
- Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer 2018;17:61. [Crossref] [PubMed]
- Zeng H, Yang Z, Xu N, et al. Connective tissue growth factor promotes temozolomide resistance in glioblastoma through TGF-beta1-dependent activation of Smad/ERK signaling. Cell Death Dis 2017;8:e2885 [Crossref] [PubMed]
- Liu B, Zhou J, Wang C, et al. LncRNA SOX2OT promotes temozolomide resistance by elevating SOX2 expression via ALKBH5-mediated epigenetic regulation in glioblastoma. Cell Death Dis 2020;11:384. [Crossref] [PubMed]
- Xu N, Liu B, Lian C, et al. Long noncoding RNA AC003092.1 promotes temozolomide chemosensitivity through miR-195/TFPI-2 signaling modulation in glioblastoma. Cell Death Dis 2018;9:1139. [Crossref] [PubMed]
- Sun L, Ma W, Gao W, et al. Propofol directly induces caspase-1-dependent macrophage pyroptosis through the NLRP3-ASC inflammasome. Cell Death Dis 2019;10:542. [Crossref] [PubMed]
- Kitazawa M, Hida S, Fujii C, et al. ASC Induces Apoptosis via Activation of Caspase-9 by Enhancing Gap Junction-Mediated Intercellular Communication. PLoS One 2017;12:e0169340 [Crossref] [PubMed]
- Knight ER, Patel EY, Flowers CA, et al. ASC deficiency suppresses proliferation and prevents medulloblastoma incidence. Oncogene 2015;34:394-402. [Crossref] [PubMed]
- Hong S, Hwang I, Lee YS, et al. Restoration of ASC expression sensitizes colorectal cancer cells to genotoxic stress-induced caspase-independent cell death. Cancer Lett 2013;331:183-91. [Crossref] [PubMed]
- Ohtsuka T, Liu XF, Koga Y, et al. Methylation-induced silencing of ASC and the effect of expressed ASC on p53-mediated chemosensitivity in colorectal cancer. Oncogene 2006;25:1807-11. [Crossref] [PubMed]
- Newton K, Wickliffe KE, Maltzman A, et al. Activity of caspase-8 determines plasticity between cell death pathways. Nature 2019;575:679-82. [Crossref] [PubMed]
- Zhao Z, Wang X, Bao XQ, et al. Autoimmune polyendocrine syndrome induced by immune checkpoint inhibitors: a systematic review. Cancer Immunol Immunother 2021;70:1527-40. [Crossref] [PubMed]
- Bates SE. Epigenetic Therapies for Cancer. N Engl J Med 2020;383:650-63. [Crossref] [PubMed]
- Parsons MJ, Patel P, Brat DJ, et al. Silencing of TMS1/ASC promotes resistance to anoikis in breast epithelial cells. Cancer Res 2009;69:1706-11. [Crossref] [PubMed]
- Hervouet E, Vallette FM, Cartron PF. Impact of the DNA methyltransferases expression on the methylation status of apoptosis-associated genes in glioblastoma multiforme. Cell Death Dis 2010;1:e8 [Crossref] [PubMed]
- Martinez R, Schackert G, Esteller M. Hypermethylation of the proapoptotic gene TMS1/ASC: prognostic importance in glioblastoma multiforme. J Neurooncol 2007;82:133-9. [Crossref] [PubMed]
- Rankeillor KL, Cairns DA, Loughrey C, et al. Methylation-specific multiplex ligation-dependent probe amplification identifies promoter methylation events associated with survival in glioblastoma. J Neurooncol 2014;117:243-51. [Crossref] [PubMed]
- Jiang Z, Li XG, Hu J, et al. The methylation and mRNA expression of SLC5A8 and TMS1/ASC genes in human glioma. Zhonghua Yi Xue Za Zhi 2007;87:292-7. [PubMed]
(English Language Editor: J. Gray)


