
Expression and prognostic analysis of Rho GTPase-activating protein 11A in lung adenocarcinoma
Introduction
Lung cancer is responsible for more mortality than any other malignancy in the world (1). Due to changes in lifestyle and increasing air pollution, lung cancer is rising in incidence every year (2). In recent years, lung adenocarcinoma (LUAD) has replaced lung squamous cell carcinoma as the most common histological type of lung cancer (3,4). With the rapid progress of medical imaging and early screening technology for lung cancer, as well as the continuous improvement of targeted lung cancer therapies, more and more lung cancer patients have benefited and significantly extended their survival. Nevertheless, at present, the 5-year survival rate of patients with non-small cell lung cancer is still less than 15% (5,6). Therefore, there is an urgent need to discover new diagnostic markers and therapeutic targets to improve the survival of patients with non-small cell lung cancer.
Rho GTPase-activating protein 11A (ARHGAP11A), also known as MP-GAP, is a member of the Rho GTPase-activating protein (RhoGAP) subfamily (7). RhoGAPs are down-regulated in tumors and are usually associated with malignant progression (8). Currently, the role of ARHGAP11A in cancer is still controversial. Recent studies have revealed that ARHGAP11A is highly expressed in colon cancer (9), basal-like breast cancer (10,11), and hepatocellular carcinoma (12). In human glioma cells, an ability of ARHGAP11A to physically bind to p53 and promote its function, to ultimately induce cell-cycle arrest and apoptosis, has been reported (13). Another study found that ARHGAP11A knockout results in G1-phase cell-cycle arrest in basal-like breast cancer cells (10). Interestingly, recent research showed that ARHGAP11A knockdown decreased cell proliferation, invasion, and migration in hepatocellular carcinoma (12,14). However, the expression and function of ARHGAP11A in LUAD has remained unclear.
Using The Cancer Genome Atlas (TCGA), Oncomine, and Kaplan-Meier (KM) plotter databases, and immunohistochemical (IHC) analysis, in this study, we apply novel and comprehensive bioinformatics methods to explored the differential expression of the ARHGAP11A gene and its correlations with clinicopathological characteristics and prognosis of LUAD, with the aim of revealing the role of ARHGAP11A in lung cancer progression and its significance as a potential prognostic biomarker. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/atm-21-2113).
Methods
TCGA expression data and clinical information
The original messenger RNA (mRNA) expression profiles of 535 LUAD tissue samples and 59 normal tissue samples were downloaded from the TCGA database (https://portal.gdc.cancer.gov/). The original detailed clinical data of 522 patients with LUAD were also downloaded from the TCGA database. The clinical information obtained included survival time and survival status, age, sex, pathological stage, and the T, N, and M stages. The clinicopathological characteristics of the patients with LUAD are detailed in Table 1.
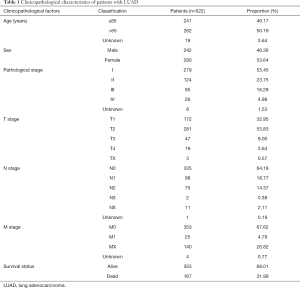
Full table
Expression difference and survival analysis of ARHGAP11A
The Perl Programming Language (version 5.32.1) was used to classify and merge the original ARHGAP11A expression data. Then, the R package limma was applied for standardization and analysis of the ARHGAP11A expression data. The limma and beeswarm packages were applied to draw scatter diagrams. Perl was applied to delete cases with incomplete survival information from the clinical data, and it was then used to combine the complete survival data and ARHGAP11A expression data. Finally, the clinical data of 494 patients with LUAD who consistent with the criterion were obtained. The LUAD cohort was divided into a high-expression group and a low-expression group on the basis of the median expression of ARHGAP11A. The R packages survival and survminer were used to perform the survival analysis and to plot KM survival curves. The survival analysis of the TCGA data was verified using the KM plotter (http://kmplot.com/) database (15), with the search condition set as: (I) cancer: lung cancer; (II) affy id/gene symbol; ARHGAP11A 204492_at; (III) split patients by: auto select best cutoff; (IV) probe set options use: only JetSet best probe set; (V) survival: overall survival (OS); (VI) follow-up threshold: all; (VII) histology: adenocarcinoma.
Verification of ARHGAP11A expression by IHC
In this study, Paraffin-embedded specimens of 54 patients with LUAD and 21 patients with normal lung tissue was collected from the Fourth Affiliated Hospital of China Medical University, and approved by the Ethics Committee of the Fourth Affiliated Hospital of China Medical University. Informed consent was taken from all the patients. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).The immunostaining assay was carried out (16) and evaluated (17) as described previously. The SP immunohistochemical kit was used in adherence with the manufacturer’s instructions. The LUAD tissue sections were incubated with the primary antibody against ARHGAP11A (1:200; NBP1-93657, Novus Biologicals) overnight at 4 °C.
Verification of ARHGAP11A expression by the Oncomine Database
The Oncomine database (http://www.oncomine.org) is an online data-mining platform that provides standardized cancer transcriptome data (18). We set the search conditions as follows: (I) gene: ARHGAP11A; (II) cancer type: LUAD; (III) analysis type: cancer vs. normal analysis; (IV) date type: mRNA; (V) data set selection threshold: P value <0.001, gene rank = ALL, fold change =1.5.
Univariate and multivariate Cox regression analysis
Perl was used to extract and merge the ARHGAP11A expression value, survival information, and clinicopathological data. Considering that the proportion of missing information for M stage exceeded 10%, the M stage was eliminated. Finally, ARHGAP11A expression, age, sex, pathological stage, T stage, and N stage were used in univariate Cox regression analysis. Factors with P<0.05 in the univariate regression analysis were subjected to multivariate Cox regression analysis. According to the median expression of ARHGAP11A as the cutoff value, the LUAD cohort was divided into the ARHGAP11A high-expression group and the ARHGAP11A low-expression group. The survival and survminer packages of R language were used to draw forest plots of the multivariate Cox regression analysis.
Gene set enrichment analysis (GSEA)
Perl was used to obtain ARHGAP11A expression value and phenotype profiles data from TCGA database. According to the median value of ARHGAP11A in the TCGA database, the LUAD cohort was divided into the ARHGAP11A high-expression group and the ARHGAP11A low-expression group. GSEA software (version 4.1.0) was used to study the potential molecular mechanism and signal transduction pathways of ARHGAP11A in LUAD (19). C2 (c2.cp.kegg.v6.2.symbols.GMT) was selected as the reference gene set for running GSEA. In GSEA, 1,000 genomic cycles are performed to identify significant pathways (20). P<0.05 and a false discovery rate (FDR) q-value served as statistical indicators.
Statistical analysis
IHC score data are expressed as mean ± standard deviation (S.D). Student’s t test was applied to analyze the expression difference of ARHGAP11A in LUAD tissues and adjacent normal tissues. Statistical analysis were performed with R software (version 4.0.3) and GraphPad Prism (version 8.0.2). P<0.05 was considered statistically significant, unless indicated otherwise.
Results
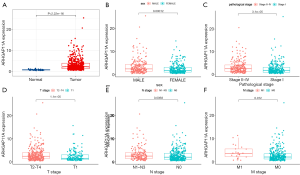
The differential expression of ARHGAP11A in LUAD
The ARHGAP11A mRNA expression data of 535 LUAD tissues and 59 adjacent normal tissues were obtained from the TCGA database. Box plots showing the difference in ARHGAP11A mRNA expression between LUAD and normal tissues can be seen in Figure 1A,B,C,D,E,F. The expression of ARHGAP11A in LUAD tissues was significantly up-regulated compared with that in normal tissues (P<0.001) (Figure 1A). Notably, the expression of ARHGAP11A in LUAD tissues was found to be closely related to sex (P<0.001, Figure 1B), pathological stage (P<0.001, Figure 1C), and advanced T stage (P<0.001, Figure 1D), N stage (P<0.01, Figure 1E), and M stage (P<0.05, Figure 1F). These results showed that the difference in ARHGAP11A expression was closely related to the malignant progression of LUAD.
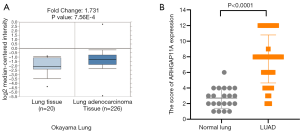
Verification of ARHGAP11A up-regulation in LUAD by Oncomine and IHC
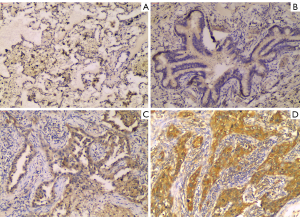
Next, we used the Oncomine database to verify the mRNA expression difference of ARHGAP11A between normal tissues and LUAD tissues, and we obtained the same result as for TCGA. As shown in Figure 2A, the mRNA expression of ARHGAP11A in LUAD tissue was significantly up-regulated in the Okatama Lung dataset (P=7.56E-4). At the same time, the protein expression difference of ARHGAP11A between normal tissues and LUAD tissues was evaluated through IHC analysis. Immunostaining of ARHGAP11A was detected in the cytoplasm. Compared with that in normal tissues, the protein expression level of ARHGAP11A in LUAD was significantly up-regulated (mean score: 7.717 vs. 2.714, P<0.0001, Figure 2B).The immunostaining of ARHGAP11A in bronchial epithelium and bronchial epithelium was generally weak (Figure 3A,B). Weak immunostaining was detected in few LUAD tissues (score <4, Figure 3C), while strong diffuse immunostaining was detected in most LUAD tissues (score ≥4, Figure 3D).
A high expression of ARHGAP11A in LUAD is related to a poor prognosis
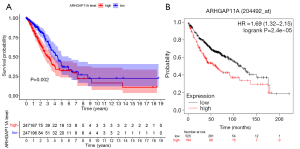
We next analyzed the prognostic significance of ARHGAP11A expression in the TCGA LUAD cohort by performing KM survival analysis. The KM survival curves showed that, compared with the ARHGAP11A low-expression cohort, the ARHGAP11A high-expression cohort showed a significant correlation with poor OS (P=0.002, Figure 4A). The 5-year overall survival rate of patients with a high expression of ARHGAP11A (32.3%) was significantly lower than that of patients with a low expression of ARHGAP11A (38.8%). The median survival of the ARHGAP11A high-expression cohort was 2.9397 years, compared with 4.3836 years in the ARHGAP11A low-expression cohort. Using the KM plotter database, we further verified that the high expression of ARHGAP11A was closely related to a poor prognosis in patients with LUAD (P=7.56E-4, Figure 4B). The median survival of the ARHGAP11A low-expression group reached 117.33 months, compared with 61.3 months in the ARHGAP11A high-expression group. The above results suggest that the high expression of ARHGAP11A is closely related to a poor prognosis of LUAD.
The relationship between ARHGAP11A expression and clinicopathological factors
To further investigate the correlation between ARHGAP11A expression and the clinicopathological features of LUAD, we obtained the clinical information of 522 patients with LUAD with complete data from the TCGA database. According to the median expression of ARHGAP11A in LUAD, the LUAD cohort was divided into a ARHGAP11A high-expression group and a ARHGAP11A low-expression group. Logistic regression analysis revealed that ARHGAP11A expression in LUAD was closely correlated with age [odds ratio (OR) =1.436, P<0.05], sex (OR =1.866, P<0.001), advanced pathological stage (P<0.05), advanced T stage (P<0.05), and lymph node metastasis (OR =1.516, P<0.05) (Table 2).

Full table
Analysis of the effect of ARHGAP11A on patient survival in LUAD by Cox proportional-hazards model
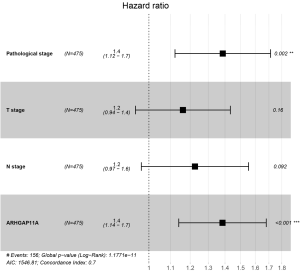
To analyze the impact of ARHGAP11A expression and other clinicopathological factors on the survival of patients with LUAD, we then applied the Cox proportional-hazards model. As the missing data for M stage exceeded 10%, Cox regression analysis was performed on 475 patients with LUAD according to sex, age, pathological stage, T stage, N stage, and ARHGAP11A expression. Univariate analysis revealed that pathological stage [hazards ratio (HR) =1.666; P<0.001], T stage (HR =1.526; P<0.001), N stage (HR =1.720; P<0.001), and ARHGAP11A expression (HR =1.516; P<0.001) could all be important predictors of survival in patients with LUAD (Table 3). Variables with P value <0.05 (ARHGAP11A expression, pathological stage, T stage, and N stage) were included in the multivariate Cox analysis. The high expression of ARHGAP11A was identified as being an independent prognostic factor for the malignant progression of LUAD (HR =1.385; P<0.001) (Figure 5 and Table 3).

Full table
Identification of ARHGAP11A-related signaling pathways in LUAD by GSEA
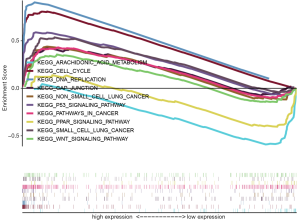
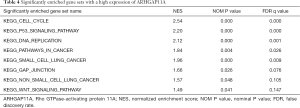
The expression and phenotype profiles of ARHGAP11A were obtained by further processing and sorting of TCGA data sets. Then, the essential functions of ARHGAP11A and its related signal transduction pathways were studied using GSEA software. According to the difference in the normalized enrichment score, nominal P value, and FDR q value, 10 significantly enriched tumor-related signaling pathways were identified. As shown in Figure 6, 8 signaling pathways involved in the cell cycle, DNA replication, gap junction, non-small cell lung cancer, P53 signaling pathway, pathways in cancer, small cell lung cancer, and WNT signaling pathways were significantly enriched in the ARHGAP11A high-expression phenotype. Meanwhile, the arachidonic acid metabolism and PPAR signaling pathways were considerably enriched in the ARHGAP11A low-expression phenotype (Figure 6 and Table 4).
Full table
Discussion
The RhoGAP family is a well-known gene family that participates in the regulation of tumor progression (21,22). Many members of the RhoGAP family have been found to be down-regulated and to have the ability to activate Rho GTPases in various cancers. Consequently, the members of this family are considered to be tumor suppressors (23). For instance, Rich1 (24) and ARHGAP29 (25) have been reported to suppress the malignant progression of cancers. ARHGAP11A has been reported to physically bind to P53 and promote its function, eventually leading to cell cycle arrest in glioma (13). However, the latest cancer research has found that ARHGAP11A is up-regulated in colon cancer, in which it promotes cell motility and invasion (9). ARHGAP11A was found to promote breast cancer invasion by inhibiting RhoA (10,11). Two previous studies found that ARHGAP11A is closely related to the malignant progression of hepatocellular carcinoma (12,14). Therefore, the expression and function of ARHGAP11A in tumors need to be clarified. Before now, the expression and mechanism of ARHGAP11A in lung cancer have remained unclear.
In this study, we explored the functional role of ARHGAP11A expression in the malignant progression and prognosis of LUAD. To clarify the potential molecular regulatory mechanism of ARHGAP11A in LUAD progression, we also screened the ARHGAP11A-related tumor signaling pathways in LUAD.
First, we analyzed the expression differences of ARHGAP11A in the TCGA database. We found that ARHGAP11A mRNA expression was significantly higher in LUAD tissues than in adjacent normal tissues. Further, Oncomine analysis and IHC assay verified that the expression of ARHGAP11A is up-regulated in LUAD. These results are consistent with the predictions of previous studies (9). We also found that the expression of ARHGAP11A was closely related to sex, advanced pathological stage, advanced T stage, N stage, and M stage. This observation suggested that ARHGAP11A expression gradually increased with the malignant progression of LUAD. Logistic regression analysis further showed that the high expression of ARHGAP11A was significantly correlated with advanced pathological stage, advanced T stage and lymph node metastasis.
KM survival analysis showed that high ARHGAP11A expression was associated with poorer OS in patients with LUAD. The survival analysis of ARHGAP11A was consistent with previous studies of lung adenocarcinoma metastasis (26). The Cox proportional-hazards model was applied to analyze the effect of ARHGAP11A on LUAD survival. The univariate Cox regression analysis showed that pathological stage, T stage, and N stage were associated with the prognosis of patients with LUAD. Multivariate Cox analysis showed that ARHGAP11A may be an independent prognostic factor of LUAD.
To explore the potential mechanism of ARHGAP11A in the malignant progression of LUAD, GSEA analysis was performed to screen ARHGAP11A-related signaling pathways. The results showed that the 10 significantly enriched signaling pathways were the arachidonic acid metabolism, cell cycle, DNA replication, gap junction, non-small cell lung cancer, P53 signaling, pathways in cancer, PPAR signaling, small cell lung cancer, and WNT signaling pathways.
In this study, we applied systematic bioinformatics methods to analyze the expression of ARHGAP11A and its clinical correlations in LUAD. This analytical approach is scientific and precise; however, our study still has some limitations. Firstly, the TCGA database does not include the specific treatment plans or surgical details of patients, which play an essential role in prognosis. However, despite this limitation, our study still supports the value of ARHGAP11A as a potential prognostic marker for LUAD. Furthermore, we will supplement the treatment information of clinical data in follow-up research and conduct an in-depth exploration of the clinical relevance of ARHGAP11A.
In conclusion, mainly through analysis of the TCGA database, our study found that ARHGAP11A is significantly up-regulated in LUAD. The expression of ARHGAP11A is closely related to clinicopathological characteristics such as sex and pathological stage. The results of our survival analysis and multivariate Cox analysis suggest that ARHGAP11A could be an independent prognostic factor in the malignant progression of LUAD. To sum up, ARHGAP11A may play a carcinogenic role in the malignant progression of LUAD, and it can be considered as a new prognostic factor and potential therapeutic target in LUAD.
Acknowledgments
Thanks to the original gene expression profile data and survival analysis provided by the TCGA database, Oncomine database, and Kaplan-Meier plotter database.
Funding: This study was supported by grants from Natural Science Foundation of Liaoning Province (No. 201602443).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-2113
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-2113
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-2113). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of the Fourth Affiliated Hospital of China Medical University. Informed consent was taken from all the patients. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Devesa SS, Bray F, Vizcaino AP, et al. International lung cancer trends by histologic type: male:female differences diminishing and adenocarcinoma rates rising. Int J Cancer 2005;117:294-9. [Crossref] [PubMed]
- Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Lee CK, Man J, Lord S, et al. Checkpoint Inhibitors in Metastatic EGFR-Mutated Non-Small Cell Lung Cancer-A Meta-Analysis. J Thorac Oncol 2017;12:403-7. [Crossref] [PubMed]
- Song Z, Ge Y, Wang C, et al. Challenges and Perspectives on the Development of Small-Molecule EGFR Inhibitors against T790M-Mediated Resistance in Non-Small-Cell Lung Cancer. J Med Chem 2016;59:6580-94. [Crossref] [PubMed]
- Zanin E, Desai A, Poser I, et al. A conserved RhoGAP limits M phase contractility and coordinates with microtubule asters to confine RhoA during cytokinesis. Dev Cell 2013;26:496-510. [Crossref] [PubMed]
- Ridley AJ. Rho proteins and cancer. Breast Cancer Res Treat 2004;84:13-9. [Crossref] [PubMed]
- Kagawa Y, Matsumoto S, Kamioka Y, et al. Cell cycle-dependent Rho GTPase activity dynamically regulates cancer cell motility and invasion in vivo. PloS One 2013;8:e83629 [Crossref] [PubMed]
- Lawson CD, Der CJ. Filling GAPs in our knowledge: ARHGAP11A and RACGAP1 act as oncogenes in basal-like breast cancers. Small GTPases 2018;9:290-6. [Crossref] [PubMed]
- Lawson CD, Fan C, Mitin N, et al. Rho GTPase Transcriptome Analysis Reveals Oncogenic Roles for Rho GTPase-Activating Proteins in Basal-like Breast Cancers. Cancer Res 2016;76:3826-37. [Crossref] [PubMed]
- Dai B, Zhang X, Shang R, et al. Blockade of ARHGAP11A reverses malignant progress via inactivating Rac1B in hepatocellular carcinoma. Cell Commun Signal 2018;16:99. [Crossref] [PubMed]
- Xu J, Zhou X, Wang J, et al. RhoGAPs attenuate cell proliferation by direct interaction with p53 tetramerization domain. Cell Rep 2013;3:1526-38. [Crossref] [PubMed]
- Lu S, Zhou J, Sun Y, et al. The noncoding RNA HOXD-AS1 is a critical regulator of the metastasis and apoptosis phenotype in human hepatocellular carcinoma. Mol Cancer 2017;16:125. [Crossref] [PubMed]
- Győrffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PloS One 2013;8:e82241 [Crossref] [PubMed]
- Guo G, Li L, Song G, et al. miR-7/SP1/TP53BP1 axis may play a pivotal role in NSCLC radiosensitivity. Oncol Rep 2020;44:2678-90. [Crossref] [PubMed]
- Li X, Tian L, Zhang L, et al. Clinical Significance of ZNF711 in Human Breast Cancer. Onco Targets Ther 2020;13:6593-601. [Crossref] [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267-73. [Crossref] [PubMed]
- Zegers MM, Friedl P. Rho GTPases in collective cell migration. Small GTPases 2014;5:e28997 [Crossref] [PubMed]
- Parri M, Chiarugi P. Rac and Rho GTPases in cancer cell motility control. Cell Commun Signal 2010;8:23. [Crossref] [PubMed]
- Csépányi-Kömi R, Sáfár D, Grósz V, et al. In silico tissue-distribution of human Rho family GTPase activating proteins. Small GTPases 2013;4:90-101. [Crossref] [PubMed]
- Zhang J, Wang J, Zhou YF, et al. Rich1 negatively regulates the epithelial cell cycle, proliferation and adhesion by CDC42/RAC1-PAK1-Erk1/2 pathway. Cell Signal 2015;27:1703-12. [Crossref] [PubMed]
- Xu Q, Duan H, Gan L, et al. MicroRNA-1291 promotes endometrial fibrosis by regulating the ArhGAP29-RhoA/ROCK1 signaling pathway in a murine model. Mol Med Rep 2017;16:4501-10. [Crossref] [PubMed]
- Li L, Peng M, Xue W, et al. Integrated analysis of dysregulated long non-coding RNAs/microRNAs/mRNAs in metastasis of lung adenocarcinoma. J Transl Med 2018;16:372. [Crossref] [PubMed]


