
Whole-exome sequencing analysis to identify novel potential pathogenetic mutations in fetuses with abnormal brain structure
Introduction
Malformations of cortical development (MCDs) encompass heterogeneous groups of structural brain anomalies which commonly cause neurodevelopmental delay and epilepsy. MCDs have been a topic of interest to clinicians and neuroscientists for many decades, as the study of these anomalies considerably advances the understanding of normal brain development and its perturbations. The majority of MCDs are believed to be attributable to underlying genetic mutations which disturb the proteins and associated signaling pathways involved in the development of the cerebral cortex (1).
Recent progress in the exploration of the genetic basis of brain malformation has been driven by advancements in high-throughput DNA sequencing technology. Mutations in different genes cause different MCDs including microcephaly (WDR62 and CDK5RAP2) (2,3), megalencephaly (AKT3 and PIK3CA) (4,5), and lissencephaly (LIS1 and TUBA1A) (6,7). Moreover, mutations in genes involved in the RTK-PI3K-AKT-mTOR pathway are known to be associated with microlissencephaly, hemimegalencephaly, and focal cortical dysplasia. Although numerous genetic causes of MCDs have been identified, other genes potentially involved in cortical development and the pathogenesis of MCDs have yet to be identified.
In the present investigation, we attempted to identify the genetic mutations associated with MCDs by whole-exome sequencing (WES) in 11 fetuses with abnormal brain structure. Dozens of candidate genes were identified as being potentially involved in cortical development. Gene ontology (GO) enrichment analysis revealed that these genes were involved in the synapse, spindle pole, centrosome, and microtubule, which was consistent with the findings of previous studies. Further, we focused on genes with mutations within multiple fetuses and predicted the protein structure variations when these mutations occurred. Our findings provide new candidate genes that are involved in cortical development and are associated with neurodevelopmental disorders.
We present the following article in accordance with the STREGA reporting checklist (available at http://dx.doi.org/10.21037/atm-21-1477).
Methods
Sample collection
Villous tissue (10–15 mg) from the placenta of pregnant women with a gestational age of 11–13+6 weeks was obtained via abdominal puncture using an 18G puncture needle (PE18/15, Italian Gallini company) under ultrasound guidance. After a gestational age of >24 weeks had been reached, 2 mL of cord venous blood was obtained via percutaneous cord blood puncture through the abdomen of each woman using a 22G puncture needle (model: B22G, Japan Hakko company) under ultrasound guidance. After the extraction of whole-genome DNA from the cord venous blood, WES was performed. This study was approved by the ethical committee of Beijing Haidian maternal and Child Health Hospital and was performed in accordance with the Helsinki Declaration (as revised in 2013). Individual consent for this retrospective analysis was waived.
Next-generation sequencing and data analysis
Sequencing was performed on an Illumina Novaseq sequencer, with a library size of 200 bp. The adaptors and low-quality reads were removed with fastp (v0.12.3, http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/) under default parameters. The remaining sequencing reads were mapped to the human reference genome (hg19, http://hgdownload.cse.ucsc.edu/goldenPath/hg19/bigZips/hg19.fa.gz) with Burrows-Wheeler Aligner (BWA, v0.7.10, http://bio-bwa.sourceforge.net) using the MEM alignment mode under default parameters. Sambamba (v0.6.8, http://lomereiter.github.io/sambamba/) was used to remove the polymerase chain reaction (PCR) duplicates.
Copy number variation identification
Each chromosome was separated into 10-kb windows, and the sequencing coverage of each 10-kb region was estimated. For reasonable comparison of the sequencing depth among different chromosomes, the base coverage of each 10-kb window was normalized by dividing it by the median coverage for the corresponding chromosome. The copy number was taken as the normalized base coverage estimated for each 10-kb region. The normalized base coverage of most genomic regions was around 1, representing 2 copies, and those of the X and Y chromosomes in male fetuses were around 0.5, representing 1 copy.
Identification of single nucleotide substitutions, insertions, and deletions
The HaplotypeCaller module in GATK (v3.8, https://github.com/broadgsa/gatk/releases) was used to detect single nucleotide substitutions (SNSs), and insertions and deletions (inDels) in each fetus under default parameters. Next, both variant types were filtered against the information in 1KGP and dbSNP130. Variants located in intergenic or intronic regions were subsequently discarded. SnpEff (v4.3t, http://snpeff.sourceforge.net/SnpEff.html#intro) was used to predict the effects of the remaining variants on genes (such as amino acid changes). Only variants with a high impact were left. Considering the randomness of the mutations, identical SNSs or inDels were unlikely to occur in more than 1 fetus. The identical genetic mutations detected in more than 2 fetuses were also discarded.
Results
No large-scale CNV was detected in any of the 11 fetuses
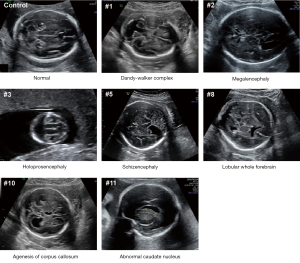
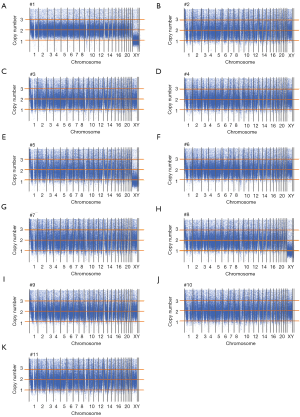
This study included 11 fetuses with abnormal brain structure (Figure 1 and Table S1). The brain abnormalities affecting the fetuses included dandy-walker complex, megalencephaly, holoprosencephaly, schizencephaly, lobular whole forebrain, agenesis of the corpus callosum, and abnormal caudate nucleus (Figure 1 and Table S1). To explore the genetic mechanisms of these abnormalities at the molecular level, umbilical blood or villus tissue samples were collected and subjected to WES. The sequencing reads were mapped to the human reference genome (hg19). To explore if there were chromosomal or genomic large-scale CNVs, the sequencing depth for each chromosome was estimated. The sequencing depth was consistent among different regions of the genome, suggesting that there were no large-scale CNVs present in the included cases (Figure 2 and Figure S1).
Genetic variants with high impact
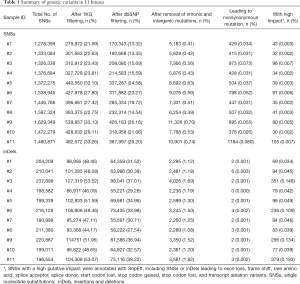
To identify the genetic variants associated with the brain structural abnormalities of the fetuses, we performed SNS calling and inDels calling using the Genome Analysis Toolkit (GATK) (8). On average, 1,441,121 SNSs and 205,966 inDels were detected per sample (Table 1). The adjacent SNSs or inDels were mainly located within 500 bp (Figures S2 and S3). To identify genetic variants associated with brain structure abnormalities, SNSs annotated in 1,000 Genome (1KGP) and dbSNP were filtered, and those within intronic and intergenic regions were removed. To further remove the genetic variants making an insignificant or no contribution to severe brain phenotypic defects, we predicted the impacts by the SNSs or inDels on genes using SnpEff (9). Finally, on average, 56 SNSs with high impact were identified per sample (Table 1), including SNSs leading to exon loss, frame shift, rare amino acid, splice acceptor, splice donor, start codon lost, stop codon gained, stop codon lost, and transcript ablation variants. Further, on average, 168 inDels with high impact were identified per sample (Table 1).
Full table
GO enrichment analysis of gene sets with high-impact mutations
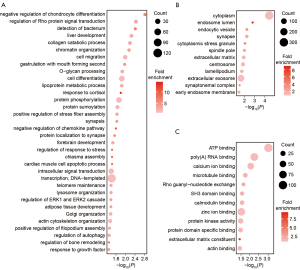
To identify the features of the SNSs and inDels identified from the samples, we studied the corresponding genes of these variants. There were 1,035 genes with high-impact genetic variants in the 11 fetuses. To explore if these genes have a common cellular function, we performed GO enrichment analysis (Table S2). Enriched biological process terms included cell migration, cell differentiation, synapsis, and forebrain development (Figure 3A). Consistently, many genes linked to the regulation of cell migration and cell differentiation are known to also be associated with abnormal neuronal migration (10-12). Cellular component enrichment analysis showed that the genes were enriched in the synapse, spindle pole, and centrosome (Figure 3B). These results are in accordance with observations by previous studies that centrosome maturation (2) and interference with mitotic spindle formation (3,13) and the spindle pole (14) affect neurogenesis, especially the cell cycle phases of mitosis. Furthermore, microtubule binding was an enriched term in molecular function GO analysis. Mutations affecting microtubule proteins have consistently been reported as being associated with abnormal neuronal migration and postmigrational development (15,16). The consistency between our results and those of previous studies demonstrate that our WES data could be used to identify genes with potential involvement in the pathogenesis of MCD. In addition to the genes known to be associated with MCDs, several other genes were identified, including a gene related to ATP binding and the regulation of Pho protein signal transduction, which proved the new genes to potentially be involved in MCD pathogenesis.
Mutations in CTDSP2
Genes with high-impact mutations detected in multiple fetuses may be more likely to be associated with brain development. We identified 7 genes with high-impact mutations in at least 3 fetuses (Table S3). These genes included CTDSP2, which is known to catalyze the dephosphorylation of ‘Ser-5’ within the tandem 7 residue repeats in the C-terminal domain (CTD) of the largest RNA polymerase II subunit POLR2A. CTDSP2 negatively regulates RNA polymerase II transcription, possibly through controlling the transition from initiation/capping to processive transcription elongation (17). CTDSP2 is also recruited by RE1-silencing transcription factor (REST) to neuronal genes containing RE-1 elements, resulting in neuronal gene silencing in non-neuronal cells (18). We retrieved the expression data of CTDSP2 from the Genotype-Tissue Expression (GTEx) project, and observed variation in the expression levels of CTDSP2 among different human brain tissues (Figure S4). Among 13 types of brain tissue, cerebellar hemisphere tissue exhibited the highest expression of CTDSP2 (Figure 4A), suggesting that mutations in CTDSP2 may lead to severe brain phenotypic defects, mainly through inaction in the cerebellar hemisphere.
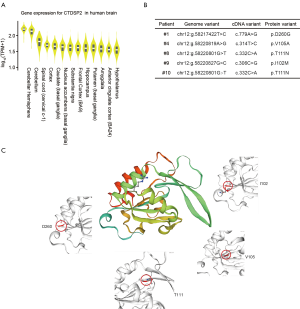
We checked 4 mutations (T111, I102, V105, and D260) in CTDSP2 in 5 fetuses carefully. All of the mutations were associated with amino acid changes in the protein sequence (Figure 4B). To understand the impact of these mutations, we predicted the protein structure. Three of these 4 mutations (I102, V105, and D260) were in the beta-pleated sheets but extremely close to the alpha-helix, which suggested that mutations in these sites may alter the protein structurally, further destroying its function (Figure 4C).
Mutations in C-terminal binding protein 2 (CTBP2)
Another gene with a high-impact mutation in 5 fetuses was the CTBP2 gene. The mammalian CTBP2 gene produces alternative transcripts encoding 2 distinct proteins, 1 of which is a transcriptional repressor (19), with the other isoform being a major component of specialized synapses known as synaptic ribbons. Both proteins contain a nicotinamide adenine dinucleotide (NAD+) binding domain similar to NAD+-dependent 2-hydroxyacid dehydrogenases. The CTBP2 expression data from GTEx showed that the expression levels of CTBP2 varied among different human brain tissues (Figure S5). Among 13 types of brain tissue, CTBP2 was also highly expressed in the cerebellar hemisphere and cerebellum (Figure 5A), indicating that mutations in CTDSP2 may cause severe brain phenotypic defects, mainly through inaction in the cerebellum.
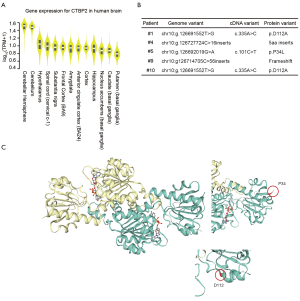
Identification of the mutation sites of the CTBP2 protein revealed that a frame shift had occurred in 2 of the 5 fetuses, while 2 nonsynonymous mutations (D112A and P34L, Figure 5B,C) had occurred in the other 3 fetuses. The predicted homodimer protein structure of CTBP2 showed that these 2 amino acid changes were outside the interaction domain, which suggested that these mutations may destroy the function of the protein by changing its structure rather than by destroying its interaction ability.
Discussion
In this study, we identified novel genetic mutations potentially associated with MCDs, and our results expand the mutation spectrum of the disease. The consistency between our observations and those of previous studies indicate the high efficiency and sensitivity of WES, as an extremely helpful approach to identifying the genetic causes of MCDs. Considering that the time and economic cost of WES have been remarkably decreased, and the interpretation of sequencing data has been largely improved, WES provides a promising application for the diagnosis of disease. Although we have provided data suggesting the potential roles of genes in the pathogenesis of MCDs, namely CTBP2, CTDSP2, HLA-DRB5, LZTFL1, MUC19, MUC4, and MUC6 (Table S3), it is necessary to discuss the limitations of our work. Firstly, to further confirm the genetic causes of MCDs the mutations in corresponding samples should be detected by Sanger sequencing. However, due to the difficulty in obtaining villous and cord venous blood from the fetuses and in order to obtain high-quality sequencing data, WES was used for all samples collected. Secondly, whether the developmental phenotype of brain tissue is abnormal should be studied in related gene knockout animal models. We intend to carry out such experiments in our future study.
The expression levels and protein structures of candidate genes should be considered in the exploration of the underlying genetic mechanisms of MCDs. In this study, we retrieved the expression data from the GTEx project and compared the expression levels of genes among different tissues. The tissues exhibiting higher expression levels were more likely to be those for which the normality of the gene should be kept. Furthermore, mutation sites and protein structural domains can together provide clues for improving the understanding of the molecular consequences of genetic mutations.
Acknowledgments
Funding: This work was supported by a grant from the Ministry of Science and Technology of the People’s Republic of China (2016YFC1000400).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-1477
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-1477
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-1477). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the ethical committee of Beijing Haidian maternal and Child Health Hospital and was performed in accordance with the Helsinki Declaration (as revised in 2013). Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Barkovich AJ, Guerrini R, Kuzniecky RI, et al. A developmental and genetic classification for malformations of cortical development: update 2012. Brain 2012;135:1348-69. [Crossref] [PubMed]
- Thornton GK, Woods CG. Primary microcephaly: do all roads lead to Rome? Trends Genet 2009;25:501-10. [Crossref] [PubMed]
- Bilgüvar K, Ozturk AK, Louvi A, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 2010;467:207-10. [Crossref] [PubMed]
- Poduri A, Evrony GD, Cai X, et al. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron 2012;74:41-8. [Crossref] [PubMed]
- Rivière JB, Mirzaa GM, O'Roak BJ, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet 2012;44:934-40. [Crossref] [PubMed]
- Reiner O, Carrozzo R, Shen Y, et al. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature 1993;364:717-21. [Crossref] [PubMed]
- Fallet-Bianco C, Loeuillet L, Poirier K, et al. Neuropathological phenotype of a distinct form of lissencephaly associated with mutations in TUBA1A. Brain 2008;131:2304-20. [Crossref] [PubMed]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491-8. [Crossref] [PubMed]
- Cingolani P, Platts A. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80-92. [Crossref] [PubMed]
- Wynshaw-Boris A. Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development. Clin Genet 2007;72:296-304. [Crossref] [PubMed]
- Ferland RJ, Batiz LF, Neal J, et al. Disruption of neural progenitors along the ventricular and subventricular zones in periventricular heterotopia. Hum Mol Genet 2009;18:497-516. [Crossref] [PubMed]
- Pramparo T, Youn YH, Yingling J, et al. Novel embryonic neuronal migration and proliferation defects in Dcx mutant mice are exacerbated by Lis1 reduction. J Neurosci 2010;30:3002-12. [Crossref] [PubMed]
- Yu TW, Mochida GH, Tischfield DJ, et al. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat Genet 2010;42:1015-20. [Crossref] [PubMed]
- Nicholas AK, Khurshid M, Desir J, et al. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat Genet 2010;42:1010-4. [Crossref] [PubMed]
- Poirier K, Keays DA, Francis F, et al. Large spectrum of lissencephaly and pachygyria phenotypes resulting from de novo missense mutations in tubulin alpha 1A (TUBA1A). Hum Mutat 2007;28:1055-64. [Crossref] [PubMed]
- Abdollahi MR, Morrison E, Sirey T, et al. Mutation of the variant alpha-tubulin TUBA8 results in polymicrogyria with optic nerve hypoplasia. Am J Hum Genet 2009;85:737-44. [Crossref] [PubMed]
- Yeo M, Lin PS, Dahmus ME, et al. A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J Biol Chem 2003;278:26078-85. [Crossref] [PubMed]
- Yeo M, Lee SK, Lee B, et al. Small CTD phosphatases function in silencing neuronal gene expression. Science 2005;307:596-600. [Crossref] [PubMed]
- Turner J, Crossley M. The CtBP family: enigmatic and enzymatic transcriptional co-repressors. Bioessays 2001;23:683-90. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


