
Long non-coding RNA X-inactive specific transcript promotes osteosarcoma metastasis via modulating microRNA-758/Rab16
Introduction
Osteosarcoma (OS) is a primary malignant bone sarcoma that originates from mesenchymal cells (1). Most OS tumors grow rapidly due to the formation of osteoid and immature bone during the cartilage stage (2). Typically, the clinical symptoms of OS include bone and joint pain, local lumps, and claudication (3). OS most commonly affects young people under the age of 20 years old (1). Due to the pathogenic complexity and highly metastatic nature of OS, its patients have unsatisfactory long-term survival (4,5). A deeper comprehension of the mechanism underpinning the growth and metastasis of OS will be of significant value for its diagnosis and therapy.
Long non-coding RNAs (lncRNAs) are critical regulatory RNAs in cells (6). Different from mRNAs, lncRNAs cannot be used as template to guide protein synthesis (6). In recent years, a large number of studies have demonstrated that lncRNAs contribute to the transcriptional and post-transcriptional modulation of multiple genes in cells (7,8). Moreover, lncRNAs are widely verified to exert tumor promoting or suppressive roles in the occurrence and development of many human cancers, including OS (9,10). LncRNA X-inactive specific transcript (XIST) is a tumor-promoting lncRNA in OS (11). Multiple studies have confirmed that XIST promotes OS progression through different pathways, while its suppression can inhibit OS progression. For instance, Sun et al. (12) reported that XIST accelerated the development of OS via the regulation of miR-375-3p/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) signaling. Further, Gao et al. (13) reported that knockdown of XIST impeded cell growth and promoted apoptosis of OS cells, and also suppressed OS xenograft tumor growth via modulating nuclear factor kappa B (NF-κB)/p53-upregulated modulator of apoptosis (PUMA) signaling. However, the other regulatory mechanisms of the effects of XIST on OS growth and metastasis demand further investigations.
MicroRNAs (miRNAs) are another type regulatory RNAs in cells, which can bind to the 3’untranslated region (3’UTR) of mRNAs, thereby silencing their translation (14). LncRNAs, including XIST, exhibit tumor-promoting or suppressive activity, usually through altering miRNA expression (15,16). The miR-758, is reported to play a tumor-suppressive role in many human cancers, including OS (17-19). In their study, Ren et al. (19) described that miR-758 limited the malignant phenotype of OS cells by regulating high mobility group 1 (HMGA1) and suppressing the Wnt/β-catenin pathway.
In the current research, we aimed to explore whether XIST serves a tumor-promoting role in OS by regulating miR-758 and to investigate possible downstream effector. The discoveries made in our research will contribute perspectives to further understanding the internal mechanism underlying the role of XIST in OS progression.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-21-1032).
Methods
Patients and tissue samples
Ten OS patients (5 female and 5 male, 29–49 years old, 2 in IIA stage, 7 in IIB stage and 1 in IIIA stage) were recruited in this study. OS tumor tissues and adjacent non-tumor tissues (from a distance of >3 cm from the tumor) were obtained during surgery and stored at –80 °C. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of Nanhua Hospital affiliated to Nanhua University. Informed consent was acquired from patients or their guardians.
Cell culture
OS cell lines (U2OS, MG-63, Saos-2, and HOS) were purchased from Procell Inc., (catalog number: CL-0236, CL-0157, CL-0202, CL-0360, Wuhan, China). Osteoblast cell line hOB was obtained from American Type Culture Collection (catalog number: CRL-11372, ATCC, VA, USA). Cells were grown in high-glucose Dulbecco’s Modified Eagle Medium (DMEM) with the addition of 10% fetal bovine serum (FBS, Sigma-Aldrich, MO, USA) and 1% penicillin-streptomycin solution (Thermo Fisher Scientific, MA, UAS). Culture flasks were placed in a humidified incubator with 5% CO2 at 37 °C.
Quantitative reverse transcription PCR (qRT-PCR) assay
Isolation of total RNA from tissues and cells was performed using TRIzol solution (Beyotime Biotechnology, Shanghai, China). The purity of the extracted RNA was tested with the BioPhotometer plus (Eppendorf, Germany). Synthesis of cDNA was carried out using PrimeScriptTM strand cDNA Synthesis Mix (TaKaRa Biomedical Technology Co., Ltd, Beijing, China). SYBR Green qPCR SuperMix (Invitrogen, CA, USA) was used to measure the relative expression levels of XIST, miR-758, and Rab16. The reaction was performed on the ABI PRISM 7500 Sequence Detection System (Applied Biosystems, CA, USA), and the reaction conditions were: 95 °C for 5 minutes, 40 cycles of 95 °C for 15 seconds, 60 °C for 32 seconds, and 72 °C for 20 seconds. GAPDH served as an endogenous control for XIST and Rab16. U6 small nuclear RNA (snRNA) served as an endogenous control for miR-758. Data were analyzed using the 2−ΔΔCt method.
Cell transfection
To form plasmids with XIST and Rab16 overexpression, the full-length sequences of XIST and Rab16 were separately inserted into pcDNA3.1 plasmid (Invitrogen), and the resulting plasmids were named pc-XIST and pc-Rab16, respectively. Small interfering RNA (siRNA) against XIST (si-XIST) was constructed by GenePharma Corporation (Shanghai, China). The miR-758 mimics (sense sequence: 5'-UUUGUGACCUGGUCCACUAACC-3', antisense sequence: 5'-UUAGUGGACCAGGUCACAAAUU-3') and miR-758 inhibitor (sequence: 5'-GGUUAGUGGACCAGGUCACAAA-3') were also obtained from GenePharma Corporation. Cell transfection was carried out using Lipofectamine 2000 Reagent in adherence with the manufacturer’s instructions.
Cell counting kit (CCK)-8 assay
A CCK-8 assay (Sigma-Aldrich) was conducted to detect the viability of MG-63 and U2OS cells. Following transfection, 1×104 MG-63 and U2OS cells were grown in a 96-well plate for 24, 48, or 72 hours. After the addition of 10 µL CCK-8 solution to the culture medium for 4 hours, the optical density (OD) value of each well was measured at 450 nm with a microplate reader (Thermo Fisher Scientific).
Cell scratch assay
After transfection, 2×105 MG-63 and U2OS cells were grown in a 6-well plate for 24 hours. A 200 µL pipette was then utilized to remove cells and create scratches. Following rinsing with phosphate-buffered saline, MG-63 and U2OS cells were cultured in free FBS-DMEM for 48 hours. The results were observed and photographed with an inverted microscope (Nikon, Japan).
Two-chamber Transwell assay
A 2-chamber Transwell assay (Costor, CA, USA) was carried out to assess MG-63 and U2OS cell migration and invasion after transfection. To study cell migration, 1×105 MG-63 and U2OS cells in 100 µL FBS-free DMEM were added into the top chamber, and the bottom chamber was filled with 700 µL DMEM containing FBS. After 48-hour culture in an incubator, a cotton swab was employed for the careful removal of the cells remaining in the top chamber. Cells that had migrated to the bottom chamber were rinsed with PBS, fixed with 4% paraformaldehyde, and dyed with violet solution. The cells were observed and photographed under a microscope. Five fields were selected, and the number of migrated cells per field was counted.
Cell invasion was assessed in a similar manner to cell migration, except that the membrane was pre-coated with Matrigel (Corning, NY, USA).
Dual luciferase reporter assay
A dual luciferase reporter assay was conducted to explore the possible binding relationship between XIST and miR-758. To form XIST wild-type (WT) reporter vector, the full-length sequence of XIST was inserted into the psiCHECK-2 plasmid (Promega, WI, USA). To form XIST mutated-type (MUT) reporter vector, the position 616-629 nucleotides of XIST was mutated and the XIST sequence containing mutated nucleotides was inserted into the psiCHECK-2 plasmid. Then, miR-758 mimics and XIST-WT reporter vector (or XIST-MUT reporter vector) were co-transfected into MG-63 cells. The ratio of luciferase activity to Renilla/Firefly activity was measured 48 hours later.
Western blotting
After transfection, total protein in MG-63 cells was collected and determined with cell lysis buffer (Beyotime Biotechnology) and a Bradford Protein Assay Kit (Beyotime Biotechnology), respectively. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out using a similar method to one described in the previous literature (20). Anti-E-cadherin antibody (ab231303), anti-N-cadherin antibody (ab76011), anti-Vimentin antibody (ab20346), and anti-GAPDH antibody (ab8245) were obtained from Abcam Biotechnology (MA, USA). Signals of proteins were visualized using an enhanced chemiluminescence technique, and relative protein expressions were analyzed with Quantity One software (Bio-Rad Laboratories, CA, USA).
Statistical analysis
All tests were repeated 3 times. Data were represented as mean ± standard deviation (SD). GraphPad Prism 6.0 (Graphpad Software, CA, USA) was employed for statistical analysis. Differences between groups (P values) were calculated using one- or two-way analysis of variance (ANOVA). P<0.05 was taken to indicate a significant difference.
Results
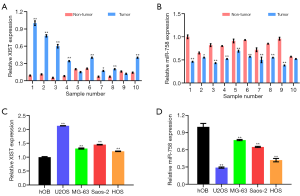
OS tissues and cells had a high expression of XIST but a low expression of miR-758
First of all, the expressions of XIST and miR-758 in OS tissues and cells were detected. Figure 1A shows that XIST was highly expressed in OS tumor tissues relative to adjacent non-tumor tissues (P<0.05 or P<0.01). Furthermore, as displayed in Figure 1B, miR-758 had a lower expression in OS tumor tissues relative to adjacent non-tumor tissues (P<0.05 or P<0.01). Similar results were found in OS cells. As illustrated in Figure 1C,D, in comparison to hOB cells, XIST was highly expressed in OS U2OS, MG-63, Saos-2, and HOS cells (P<0.01), whereas miR-758 was expressed at a low level in OS U2OS, MG-63, Saos-2, and HOS cells (P<0.01). These outcomes implied that XIST might serve an oncogenic role in OS, whereas miR-758 might engage in tumor-suppressive activity.
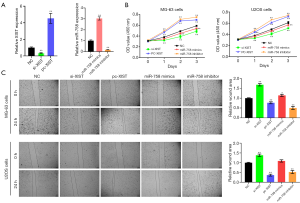
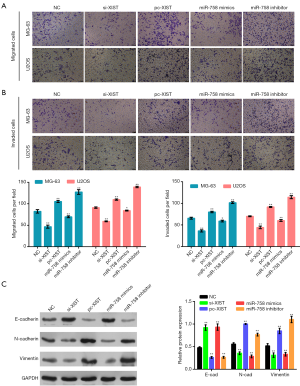
XIST knockdown and miR-758 mimics suppressed OS cell migration, invasion, and epithelial-mesenchymal transformation (EMT), while XIST overexpression and miR-758 inhibitor produced the opposite outcomes
MG-63 and U2OS cells were selected for the subsequent experiments. To knockdown and overexpress XIST, cells were transfected with si-XIST and pc-XIST, respectively. For the alteration of miR-758 expression, miR-758 mimics and miR-758 inhibitor were also transfected into cells. Results presented in Figure 2A reveal that si-XIST transfection notably lowered the XIST expression in MG-63 cells (P<0.01), while pc-XIST transfection raised the XIST expression in MG-63 cells (P<0.01). Moreover, miR-758 mimics enhanced the miR-758 expression in MG-63 cells (P<0.01), but miR-758 inhibitor had opposite influence (P<0.01). Similar results were displayed in U2OS cells (data not shown). Figure 2B showed that si-XIST or miR-758 mimics transfection reduced the viability of MG-63 and U2OS cells (P<0.05 or P<0.01). In contrast, a rise in the viability of MG-63 and U2OS cells was observed following transfection with pc-XIST or miR-758 inhibitor (P<0.01). Cell scratch assay, the results of which are displayed in Figure 2C, demonstrated that transfection with si-XIST or miR-758 mimics suppressed the migratory abilities of MG-63 and U2OS cells (P<0.05 or P<0.01), while transfection with pc-XIST or miR-758 inhibitor produced the opposite outcome (P<0.01). Also, as Figure 3A illustrates, MG-63 and U2OS cell migration was decreased after transfection with si-XIST or miR-758 mimics (P<0.05 or P<0.01), but was increased after transfection with pc-XIST or miR-758 inhibitor (P<0.01). As displayed in Figure 3B, transfection with si-XIST or miR-758 mimics inhibited the invasion ability of both MG-63 and U2OS cells, as evidenced by the decreased numbers of invading MG-63 and U2OS cells in the si-XIST and miR-758 mimics groups compared to the NC group (P<0.01). Meanwhile, transfection with pc-XIST or miR-758 inhibitor had the opposite effect, enhancing the invasion of MG-63 and U2OS cells (P<0.01). Besides, the results of western blot showed that transfection with si-XIST or miR-758 mimics up-regulated E-cadherin at the protein level (P<0.01) while down-regulated the protein levels of N-cadherin and vimentin (P<0.01), in both MG-63 and U2OS cells (Figure 3C). Inversely, transfection with pc-XIST or miR-758 inhibitor produced the opposite outcomes, with down-regulation of the level of E-cadherin protein (P<0.01) and up-regulation of the protein levels of N-cadherin and vimentin (P<0.01) observed in both MG-63 and U2OS cells. The above outcomes suggested that XIST could promote OS cell migration, invasion, and EMT, while miR-758 could inhibit OS cell migration, invasion, and EMT.
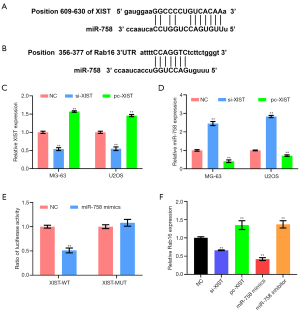
XIST regulated miR-758 expression in OS cells, while miR-758 regulated Rab16 expression
Next, the possible regulatory effects of XIST on miR-758, as well as those of miR-758 on Rab16 were explored. Bioinformatics analyses found that miR-758 could reversely bound to the sequences of XIST and Rab16 3’UTR (Figure 4A,B). Figure 4C shows that XIST expression in both MG-63 and U2OS cells was markedly decreased after si-XIST transfection (P<0.01) but significantly increased after pc-XIST transfection (P<0.01). As shown by Figure 4D, si-XIST transfection markedly raised miR-758 expression in MG-63 and U2OS cells (P<0.01), whereas pc-XIST transfection had the reverse effect, with its reduction of miR-758 expression in MG-63 and U2OS cells (P<0.01). The results of dual-luciferase activity displays that co-transfection of XIST-WT and miR-758 mimics notably reduced the ratio of luciferase activity (Figure 4E, P<0.01), while miR-758 mimics and XIST-MUT co-transfection had no significant influence on luciferase activity. Furthermore, the results in Figure 4F show that si-XIST and miR-758 mimics both reduced the expression of Rab16 in MG-63 cells (P<0.01), whereas pc-XIST or miR-758 inhibitor enhanced Rab16 expression (P<0.01). These outcomes suggested that XIST regulated miR-758 expression, and miR-758 regulated Rab16 expression in OS cells.
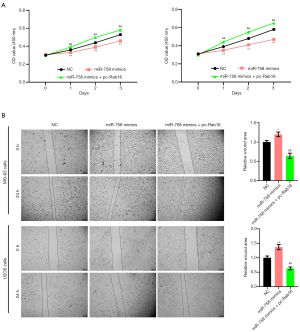
miR-758 mimics suppressed OS cell migration and invasion by down-regulating Rab16
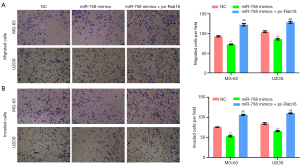
Our final experiment had the aim of investigating whether miR-758 participated in the regulation of OS cell migration and invasion via modulating Rab16. Elevation of Rab16 expression was achieved by transfecting MG-63 and U2OS cells with pc-Rab16. As evidenced in Figure 5A, pc-Rab16 transfection significantly reversed the miR-758 mimic-induced decreases of MG-63 and U2OS cell viability (P<0.01). Furthermore, relative to the miR-758 mimics group, the MG-63 and U2OS cells in the miR-758 mimics+pc-Rab16 group showed an increase in migration, which is illustrated in Figure 5B (P<0.01). Figure 6A also shows that pc-Rab16 transfection reversed the decreases of MG-63 and U2OS cell migration caused by miR-758 mimics (P<0.01), which was evidenced by an increased number of migrated MG-63 and U2OS cells in the miR-758 mimics+pc-Rab16 group compared to the miR-758 mimics group. Besides, as presented in Figure 6B, pc-Rab16 transfection also reversed the decreases of MG-63 and U2OS cell invasion observed following miR-758 mimic transfection (P<0.01). These outcomes suggested that miR-758 mimics suppressed OS cell migration and invasion through down-regulating Rab16.
Discussion
OS not only threatens patients’ lives, but also brings tremendous mental and economic pressure to the patients and their families (4). Tumor recurrence and metastasis are the main reasons for the death of patients who have OS (21). In recent years, an increasing number of non-coding RNAs, particularly lncRNAs and miRNAs, have been verified to play key roles in the occurrence and development of OS (16). It is a main aim of researchers to gain an understanding of the molecular mechanisms of OS occurrence and development in relation to non-coding RNAs, and to find suitable lncRNAs or miRNAs to serve as targets for OS therapy. Since lncRNA XIST was first discovered in 1992 (22), many studies have confirmed its oncogenic role in human cancers, such as pancreatic cancer (23), lung adenocarcinoma (24), and cervical cancer (25). More importantly, from our perspective, several literatures have reported the tumor-promoting activity of XIST in OS (12,13,26,27). In the current research, we further confirmed that XIST was highly expressed in OS tissues and cells, and acted as a promoter of OS cell migration and invasion. XIST knockdown could inhibit the migration and invasion abilities of OS cells. EMT is believed to be essential for tumor metastasis, which endows tumor cells with migratory and invasive abilities (28). As key EMT proteins, E-cadherin expression was decreased, while the expressions of N-cadherin and vimentin were increased, during EMT (28). We discovered that XIST overexpression lowered E-cadherin expression, but elevated the levels of N-cadherin and vimentin in OS cells. Meanwhile, XIST knockdown had the reverse effect. These findings were consistent with the results of Wen et al.’s research (29), and further implied that XIST could be used as potential target for OS therapy, although more in vivo and clinical researches are needed to investigate this.
Previous literatures reported that XIST performed an oncogenic role in OS via modulating miR-153 (29), miR-375-3p (12), miR-195-5p (26), miR-320b (30), miR-137 (31), and miR-193-3p (32). miR-758 is involved in tumor inhibition in multiple human cancers and also takes part in suppressing tumor metastasis (17,33,34). Wu et al. (34) found that miR-758-3p restricted the proliferation, migration, and invasion of bladder cancer cells, and Song et al. (35) reported that miR-758 suppressed cell growth and metastasis in cervical cancer. Moreover, miR-758 was observed to be negatively related to tumor size, clinical stage, and distant metastasis in patients with OS (19). In this research, we revealed that, different from XIST, miR-758 was expressed at low levels in OS tissues and cells. Also, miR-758 mimics inhibited the migration, invasion, and EMT of OS cells, while miR-758 inhibitor had the opposite effects. Additionally, we discovered that XIST could bind and negatively modulate miR-758 expression in OS cells, which hinted that miR-758 might be a downstream miRNA of XIST. XIST contributed to OS cell growth and metastasis might be achieved via modulating miR-758 expression. Considering all of the previous findings, we propose that there are multiple miRNAs involved in the oncogenic role of XIST in OS, and together, they may constitute a complex regulatory network.
Rab proteins are one type of GTPases, which is related to the transport of intracellular proteins in various organelles (36). So far, more than 60 different Rab proteins have been found in cells (36). Some Rab proteins, including Rab7A (37), Rab22A (38), Rab27 (39), and Rab31 (40), have been shown to be expressed at higher levels in OS tissues and cells. Moreover, all of the above-mentioned Rabs are modulated by miRNAs in OS cells. Herein, we found that Rab16 was positively regulated by XIST in OS cells but negatively modulated by miR-758. More importantly, overexpression of Rab16 reversed the effects of miR-758 mimics on OS cell migration and invasion. These findings suggest that Rab16 was a downstream effector of XIST/miR-758 in OS cells. Considering that miR-758 had been discovered to directly targeting modulate high mobility group AT-hook 1 (HMGA1) in OS (19), we proposed that Rab16 was not the only downstream target of miR-758 in OS. Further investigations are still needed.
In conclusion, this research further affirmed the tumor-promoting role of XIST in OS progression. XIST contributes to OS cell migration, invasion, and EMT through its modulation of miR-758/Rab16. Therefore, XIST might be a suitable lncRNA target for OS therapy.
Acknowledgments
Funding: This work was supported by Hunan Education Department Project (No. 19C1567) and Clinical Medical Technology Innovation Guiding Project of China Hunan Provincial Science and Technology Department (No. 2020SK51902).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-1032
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-1032
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-1032). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of Nanhua Hospital affiliated to Nanhua University. Informed consent was acquired from patients or their guardians.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Biazzo A, De Paolis M. Multidisciplinary approach to osteosarcoma. Acta Orthopaedica Belgica 2016;82:690-8. [PubMed]
- Picci P. Osteosarcoma (Osteogenic sarcoma). Orphanet J Rare Dis 2007;2:6. [Crossref] [PubMed]
- Widhe B, Widhe T. Initial Symptoms and Clinical Features in Osteosarcoma and Ewing Sarcoma. J Bone Joint Surg Am 2000;82:667-74. [Crossref] [PubMed]
- Durfee RA, Mohammed M, Luu HH. Review of Osteosarcoma and Current Management. Rheumatol Ther 2016;3:221-43. [Crossref] [PubMed]
- Heymann MF, Lézot F, Heymann D. The contribution of immune infiltrates and the local microenvironment in the pathogenesis of osteosarcoma. Cell Immunol 2019;343:103711 [Crossref] [PubMed]
- Chen H, Shan G. The physiological function of long-noncoding RNAs. Noncoding RNA Res 2020;5:178-84. [Crossref] [PubMed]
- Moore JB, Uchida S. Functional characterization of long noncoding RNAs. Curr Opin Cardiol 2020;35:199-206. [Crossref] [PubMed]
- Xu Z, Yan Y, Qian L, et al. Long non-coding RNAs act as regulators of cell autophagy in diseases Oncol Rep 2017;37:1359-66. (Review). [Crossref] [PubMed]
- Zhang Y, Pu Y, Wang J, et al. Research progress regarding the role of long non-coding RNAs in osteosarcoma Oncol Lett 2020;20:2606-12. (Review). [Crossref] [PubMed]
- Guzel E, Okyay TM, Yalcinkaya B, et al. Tumor suppressor and oncogenic role of long non-coding RNAs in cancer. North Clin Istanb 2019;7:81-6. [Crossref] [PubMed]
- Wang W, Shen H, Cao G, et al. Long non-coding RNA XIST predicts poor prognosis and promotes malignant phenotypes in osteosarcoma. Oncol Lett 2019;17:256-62. [PubMed]
- Sun X, Wei B, Peng ZH, et al. Knockdown of lncRNA XIST suppresses osteosarcoma progression by inactivating AKT/mTOR signaling pathway by sponging miR-375-3p. Int J Clin Exp Pathol 2019;12:1507-17. [PubMed]
- Gao W, Gao J, Chen L, et al. Targeting XIST induced apoptosis of human osteosarcoma cells by activation of NF-kB/PUMA signal. Bioengineered 2019;10:261-70. [Crossref] [PubMed]
- Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol 2018;141:1202-7. [Crossref] [PubMed]
- Zhang R, Xia T. Long non-coding RNA XIST regulates PDCD4 expression by interacting with MIR-21-5p and inhibits osteosarcoma cell growth and metastasis. Int J Oncol 2017;51:1460-70. [Crossref] [PubMed]
- Wang JY, Yang Y, Ma Y, et al. Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed Pharmacother 2020;121:109627 [Crossref] [PubMed]
- Liu J, Jiang J, Hui X, et al. Mir-758-5p Suppresses Glioblastoma Proliferation, Migration and Invasion by Targeting ZBTB20. Cell Physiol Biochem 2018;48:2074-83. [Crossref] [PubMed]
- Chen J, Xu Z, Yu C, et al. MiR-758-3p regulates papillary thyroid cancer cell proliferation and migration by targeting TAB1. Die Pharmazie 2019;74:235-8. [PubMed]
- Ren J, Yang M, Xu F, et al. microRNA-758 inhibits the malignant phenotype of osteosarcoma cells by directly targeting HMGA1 and deactivating the Wnt/β-catenin pathway. Am J Cancer Res 2019;9:36-52. [PubMed]
- Li R, Yin F, Guo YY, et al. Knockdown of ANRIL aggravates H2O2-induced injury in PC-12 cells by targeting microRNA-125a. Biomed Pharmacother 2017;92:952-61. [Crossref] [PubMed]
- Endo-Munoz L, Evdokiou A, Saunders N. The role of osteoclasts and tumour-associated macrophages in osteosarcoma metastasis. Biochimica Et Biophysica Acta 2012;1826:434-42. [PubMed]
- Brown CJ, Hendrich BD, Rupert JL, et al. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 1992;71:527-42. [Crossref] [PubMed]
- Wei W, Liu Y, Lu Y, et al. LncRNA XIST Promotes Pancreatic Cancer Proliferation through miR-133a/EGFR. J Cell Biochem 2017;118:3349-58. [Crossref] [PubMed]
- Sun J, Pan LM, Chen LB, et al. LncRNA XIST promotes human lung adenocarcinoma cells to cisplatin resistance via let-7i/BAG-1 axis. Cell Cycle 2017;16:2100-7. [Crossref] [PubMed]
- Zhu H, Zheng T, Yu J, et al. LncRNA XIST accelerates cervical cancer progression via upregulating Fus through competitively binding with miR-200a. Biomed Pharmacother 2018;105:789-97. [Crossref] [PubMed]
- Yang C, Wu K, Wang S, et al. XIST Promotes Osteosarcoma Progression via miR-195-5p/YAP axis. J Cell Biochem 2018;119:5646-56. [Crossref] [PubMed]
- Li GL, Wu YX, Li YM, et al. High expression of long non-coding RNA XIST in osteosarcoma is associated with cell proliferation and poor prognosis. Eur Rev Med Pharmacol Sci 2017;21:2829-34. [PubMed]
- Du L, Tang JH, Huang GH, et al. The progression of epithelial-mesenchymal transformation in gliomas. Chinese Neurosurgical Journal 2017;3.
- Wen JF, Jiang YQ, Li C, et al. LncRNA-XIST promotes the oxidative stress-induced migration, invasion and epithelial to mesenchymal transition of osteosarcoma cancer cells through miR-153-SNAI1 axis. Cell Biol Int 2020;44:1991-2001. [Crossref] [PubMed]
- Lv GY, Miao J, Zhang XL. Long Noncoding RNA XIST promotes Osteosarcoma progression by targeting Ras-Related protein RAP2B via miR-320b. Oncol Res 2018;26:837-46. [Crossref] [PubMed]
- Li H, Cui J, Xu B, et al. Long non-coding RNA XIST serves an oncogenic role in osteosarcoma by sponging miR-137. Exp Ther Med 2019;17:730-8. [PubMed]
- Wu D, Nie X, Ma C, et al. RSF1 functions as an oncogene in osteosarcoma and is regulated by XIST/miR-193a-3p axis. Biomed Pharmacother 2017;95:207-14. [Crossref] [PubMed]
- Jiang D, Cho W, Li Z, et al. MiR-758-3p suppresses proliferation, migration and invasion of hepatocellular carcinoma cells via targeting MDM2 and mTOR. Biomed Pharmacother 2017;96:535-44. [Crossref] [PubMed]
- Wu X, Chen B, Shi H, et al. miR-758-3p suppresses human bladder cancer cell proliferation, migration and invasion by targeting NOTCH2. Exp Ther Med 2019;17:4273-8. [PubMed]
- Song T, Hou X, Lin B. MicroRNA-758 inhibits cervical cancer cell proliferation and metastasis by targeting HMGB3 through the WNT/β-catenin signaling pathway. Oncol Lett 2019;18:1786-92. [Crossref] [PubMed]
- Lamber EP, Siedenburg AC, Barr FA. Rab regulation by GEFs and GAPs during membrane traffic. Curr Opin Cell Biol 2019;59:34-9. [Crossref] [PubMed]
- Zhang YY, Xu HY, Dai JJ. MiR-193b enhanced proliferation and migration and inhibits apoptosis through targeting RAB7A in osteosarcoma cell. Mol Cell Toxicol 2021;17:69-78. [Crossref]
- Zheng S, Jiang F, Ge D, et al. LncRNA SNHG3/miRNA-151a-3p/RAB22A axis regulates invasion and migration of osteosarcoma. Biomed Pharmacother 2019;112:108695 [Crossref] [PubMed]
- Pu Y, Zhao F, Cai W, et al. MiR-193a-3p and miR-193a-5p suppress the metastasis of human osteosarcoma cells by down-regulating Rab27B and SRR, respectively. Clin Exp Metastasis 2016;33:359-72. [Crossref] [PubMed]
- Wu Q, Feng Q, Xiong Y, et al. RAB31 is targeted by miR-26b and serves a role in the promotion of osteosarcoma. Oncol Lett 2020;20:244. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


