
CTNNB1 S37C mutation causing cells proliferation and migration coupled with molecular mechanisms in lung adenocarcinoma
Introduction
Lung adenocarcinoma (LUAD) is a kind of malignant tumor originated from the glandular epithelium of bronchial mucosa, accounting for approximately 40% of lung cancer (1-3). In recent years, around 1.8 million people worldwide die of fatal diseases caused by LUAD every year (3). At present, the main treatment methods are surgery, radiotherapy and chemotherapy (4-6). Smoking is responsible for primary cause of LUAD. However, as smoking rates have declined, proportionately more cases have occurred among never-smokers (7,8). Even worse, the incidence of LUAD has been increasing worldwide in the past few decades (9).
The mutation of certain gene is demonstrated to be implicated in the incidence and development of LUAD (10). Catenin beta 1 (CTNNB1), a protein encoding by CTNNB1 gene, has been reported to play roles in regulating genes transcription, cell adhesion and other cellular processes (11). CTNNB1 mutation has been found in various cancers (12,13). The β-catenin protein is a key effector in Wnt/β-catenin signaling pathway during the development of human cancer (14). The aggregation of β-catenin protein in the cytoplasm induced by CTNNB1 gene mutation can translocate into the nucleus and result in abnormal expression in cells, which further cause the abnormal activating of Wnt/β-catenin signaling pathway (15,16). Previous studies indicate that β-catenin-TCF/LEF interaction in cytoplasm can promote the transcription of downstream target genes and causes abnormal proliferation and differentiation of cells, thus promoting the occurrence of cancer (17,18). Actually, the site mutation of CTNNB1 always associate with the changes of CTNNB1 biological function in various humane diseases (19-21). A previous study shows that the CTNNB1 mutations trigger the high expression of Wnt signaling pathway, which further contribute to the development of LUAD (22). Importantly, CTNNB1 mutation suggests a poor prognosis the prognostic in LUAD (23). Our recent clinicopathologic characteristics investigation of LUAD with CTNNB1 mutation showed that women and non-smokers may have CTNNB1 mutation, and the LUAD patients who have CTNNB1 mutation tended to worse outcomes (24). However, the potential mechanism of the effect of CTNNB1 mutation on LUAD progression are still unclear.
In this study, LUAD cell lines with CTNNB1 S37C mutation were established. Cells proliferation and migration were promoted in LUAD cells with CTNNB1 S37C mutation. We further performed transcriptome sequencing to investigate the potential molecular mechanism, and large-scale gene expression changes were found in LUAD cells with CTNNB1 S37C mutation. Our study demonstrates the cytological effects of CTNNB1 S37C mutation in LUAD, and provide potential targets to investigate the molecular mechanisms.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-21-1146).
Methods
Cell culture
LUAD NCI-H1299 cells and NCI-H1975 cells (Cell Bank, Chinese Academy of Sciences, Shanghai) were maintained in RPMI 1640 culture medium (Gibco BRL, catalog No. C11875500BT) containing 10% fetal bovine serum (Gibco BRL, catalog No. 10099141) and 1% penicillin/streptomycin (Gibco BRL, catalog No. 15140-122), followed by incubation at 37 °C with 5% CO2.
Vector construction and cell transfection
The S37C mutation (TCT to TGT) of CTNNB1 gene (total length: 2346 bp) was established, followed by the pCDH-CMV-MCS-EF1-puro (CD510B) vector construction via XbaI/NotI restriction site. After plasmid extraction, digestion and agarose gel electrophoresis, the plasmids with matching band sizes were sequenced (forward 1: GCACCAAAATCAACGGGACT; forward 2: GCAGAAAATGGTTGCCTTGC; forward 3: GCACATCAGGATACCCAGCG) by Sangon Biology (Shanghai) Technology Co., Ltd. Then, plasmids from the 100 mL LB culture medium were extracted using Plasmid Extraction Kit (Tiangen, catalog No. DP116), and used for the following analysis. Three groups were set during cell transfection, including normal control (NC) group, wide type (WT) group and mutation group. Cells in NC and WT groups were transfected with empty vector, and CTNNB1, respectively. While cells in mutation group were transfected with CTNNB1 mutation. Lipfectamine 2000 (Invirtrogn, catalog No. 11668019) was used to complete the cell transfection in accordance with the manufacturer’s instructions.
Real-time quantitative PCR
The extraction of total RNA was completed by Trizol (TaKaRa, catalog No. 9109), followed by quantitation utilizing RNAiso Plus. The cDNA template was synthesized utilizing 5× primeScript RT Master MIX (TaKaRa, catalog No. RR036A) in accordance with the instructions. Then, a total of 2 µL cDNA was used for PCR investigation. The reaction conditions were as follows: 98 °C for 5 min, 30 cycles at 98 °C for 30 s, 55 °C for 30 s, 68 °C for 30 s, as well as 68 °C for 10 min. Relative expression of candidate genes were calculated by 2−∆∆Ct method (25). The primer sequences used in this assay were recorded in Table 1.
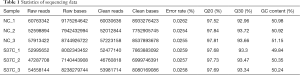
Full table
Western blot
Total proteins of samples in each group were extracted by RIPA lysis buffer (Beyotime Biotechnological Co., Ltd, catalog No. P0013B). After centrifugation, proteins were separated by 10% SDS-polyacrylamide gel electrophoresis, and performed with the polyvinylidene fluoride membrane (Millipore). Then the membrane was blocked with 5% skim milk (0.75 g milk powder + 15 mL PBS) for 1 h, and incubated with primary antibodies (β-catenin, rabbit monoclonal antibody, 1:1,000, SCT, catalog No. 8480S; GAPDH, rat monoclonal antibody, 1:1,000, Santa Cruz; catalog No. sc-47724) overnight at 4 °C. Then, samples were treated with the secondary antibody (anti-rabbit, 1:10,000; anti-rat, 1:5,000; Jackson) for 2 h at 37 °C. Protein brands were visualized using Millipore ECL Gel imaging system (Millipore, Billerica, MA, USA). Finally, the results were analyzed by TanonImage 4600 (Tanon).
CCK-8 assay
A total of 5 mg/mL CCK solution (Beyotime Biotechnological Co., Ltd, catalog No. C0040) was used for the CCK-8 assay on 1 mL 0.25% trypsin (Gibco, catalog No. 25200-072) digested cells (transfected for 24 hours). All cells were inoculated into the 96-well plate (6×103 cells/well, 100 µL/well) at 37 °C with 5% CO2. Then, the 1640 medium containing 10% CCK-8 solution (5 mg/mL) was added into the 96-well plate at 24, 48, 72 and 96 h after incubation. Finally, the 450 nm absorbance for each was recorded by Infinite M100 PRO microplate reader (TECAN, Gene Co., Ltd.) after treatment of CCK8 solution for 1 h.
Scratch assay
After adjusting the cell density of each group, a total of 1 mL 0.25% trypsin (Gibco, catalog No. 25200-072) digested cells (transfected for 24 hours) were seeded into the 12-well plate (4×105/cells). Following drew a line on the culture medium surface, washed using PBS and supplemented with fresh culture medium. Then cells were maintained for 24 h (37 °C with 5% CO2). Finally, cell migration rate was calculated by observing cells and photographing cells on an inverted microscope (Olympus Ckx53).
Transcriptome sequencing
NC group (n=3) and mutation group (n=3) in H1299 cells were used for transcriptome sequencing analysis after 48 hours of transfection. Briefly, total RNA was extracted and examined by spectrophotometer. Moreover, NEBNext® Ultra™ DNA Library Prep Kit for Illumina® (New England Biolabs, Inc., Ipswich, MA) was used to construct the RNA-seq library. Finally, the sequencing data were collected using the Illumina Hiseq 4000 platform (PE150) (Illumina, San Diego, CA, USA). The sequencing results were preserved for subsequent analysis
Quality control and expression analysis
SeqPrep, Sickle and HISAT2software were implemented to sort raw sequencing data. Reads was assembled using StringTie (Version 1.3.3b). Furthermore, the RSeQC (version: 2.3.6) software was used for the transcriptome quality assessment. Principal component analysis was performed to assess the correlations among samples.
The differential expression analysis of DEGs
The reads counts were analyzed with DESseq2 software based on negative binomial distribution to reveal the genes differentially expressed between two groups. Benjamini/Hochberg method was used to perform FDR correction. The adjusted P value <0.05 and |log2 fold change (FC)| ≥1 were used as cut-off values for DEGs analysis. The results were visualized by heatmap and volcano plot.
The functional and pathway enrichment analysis
Gene ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analysis for DEGs were performed based on Goatools software. Bonferroni corrected P<0.05 was considered as cut-off values for GO function investigation. Meanwhile, BH corrected P<0.05 was considered as cut-off values for KEGG pathway investigation. Finally, the results were visualized by bar-chart.
PPI network construction
Based on the STRING database (version: 11) (26), the interactions between DEGs were predicted. The interactions among proteins that encoded by the co-DEGs were selected according to STRING database with medium confidence (score) >0.4, followed by the PPI network construction using Cytoscape. (version: 3.7.2) (27).
Statistical analysis
Statistical analyses were using prism 8 software (GraphPad Software, USA). All values were presented as means ± standard deviation (SD). One-way ANOVA was used for comparison between groups, followed by Newman-Keuls test. P<0.05 was selected to present statistically significant.
Results
Overexpressing CTNNB1 inhibited cell proliferation and migration in LUAD cells
The successful establishment of CTNNB1 mutation was tested by PCR assay. After transfection, results of western blot indicated that the expression of the β-catenin increased in both NCI-H1299 and NCI-H1975 cells (Figure 1A). These results indicated that CTNNB1 with S37C mutation were successfully expressed in LUAD cells, and CTNNB1 S37C mutation seemed to activate the expression of β-catenin
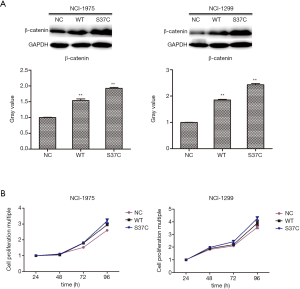
We further performed CCK-8 assay to investigate the cell proliferation in two LUAD cells. The results indicated that cell proliferation of H1975 cells and H1299 cells showed an increased trend in a time-dependent manner (Figure 1B). These results indicated that CTNNB1 S37C mutation contributed to the cell proliferation in both H1975 and H1299 LUAD cell lines.
Cell migration was promoted by CTNNB1 S37C mutation
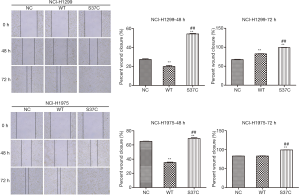
The effect of CTNNB1 S37C mutation on the cell migration was determined utilizing cell scratch assay. Compared with NC and WT group, the migration rate of H1299 cells in CTNNB1 S37C mutation group was significantly elevated at 48 h and 72 h (P<0.01), and similar results were observed in H1975 cells (Figure 2). This suggested that CTNNB1 S37C mutation promoted the migratory property of LUAD cells. However, the migration rate of both H1299 and H1975 cells in WT group were significantly decreased at 48 h in comparison with that of NC group. While the migration rate of H1299 in WT group were significantly increased at 72 h in comparison with that of NC group. The migration rate of H1975 at 72 h showed no significant difference between WT and NC group (Figure 2).
Sequencing data statistics and DEG screening
The statistics of raw reads and clean reads were shown in Table 1. A total of 169,266,638 clean reads were obtained for samples in NC group, while 153,219,672 clean reads were obtained for samples in CTNNB1 S37C mutation group. After quality control analysis and data preprocessing, a total of 13531 genes were screened from the all the 6 samples (Figure S1A). The PCA analysis for samples showed that there were significant differences for the samples in the two groups (Figure S1B).
DEGs investigation of CTNNB1 mutation
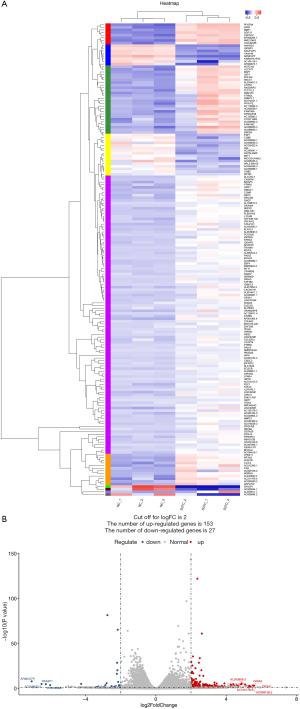
With |logFC| >2 and P<0.05, a total of 1060 DEGs including 153 up-regulated DEGs and 27 down-regulated DEGs were revealed between NC and CTNNB1 mutation groups. The heatmap and volcano plot for genes in two groups were showed in Figure 3. These results showed that samples could be well distinguished by DEGs.
The enrichment analysis of DEGs
The GO function analysis was performed for DEGs between CTNNB1 mutation and NC groups. The result showed that these DEGs were mainly assembled in functions like extracellular matrix (GO:0030198), extracellular space (GO:0005615) and cell adhesion (GO:0007155). The top 20 of GO functions assembled by DEGs were showed in Figure 4A. Moreover, these DEGs were mainly enriched in pathways including nicotine addiction (map05033), MAPK signaling pathway (MAPK signaling pathway) and gap junction (map04540) (Figure 4B).
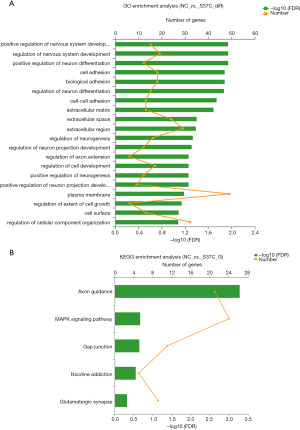
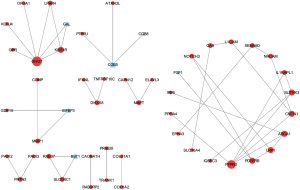
PPI network investigation
A PPI network was constructed based on the 180 DEGs corresponding proteins and associated interactions. The result showed that there were 51 DEGs and 45 interactions in PPI network. The detail information for PPI network was showed in Figure 5. PTPRD, GNG7 and CNTN1 were hub genes in PPI network with higher degree. CGB5 interacted with PTPRU, while IGFBP3 showed interaction with MMP1.
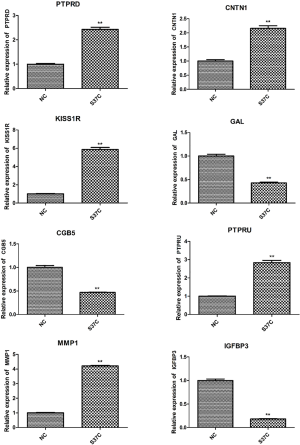
The qRT-PCR verification analysis for DEGs
We next selected and verified expression of several DEGs by using qRT-PCR (Figure 6). It could be seen that the expression of PTPRD, CNTN1, KISS1R, PTPRU and MMP1 were significantly elevated (P<0.01) in CTNNB1 S37C mutation group in comparison with that of control group. Dissimilarly, the expression of GAL, CGB5 and IGFBP3 were significantly decreased (P<0.01) in CTNNB1 S37C mutation group. These results were consistent with the results of sequencing (Figure S2).
Discussion
LUAD is considered as one of the main causes of cancer death worldwide. Although mutations of CTNNB1 have been found and reported in various human cancers, including LUAD (24), the effect of CTNNB1 mutation on LUAD mechanism is still unclear. CTNNB1 is a proto oncogene encoding β-catenin protein, which can cause multiple cancers after mutation (28,29). The β-catenin serves as an crucial component of Wnt/β-catenin pathway, and has been demonstrated to up-regulate in the development of a broad range of tumors (30,31). A previous study indicates that the β-catenin encoded by CTNNB1 increases proliferative and migratory abilities of cells, and triggers apoptosis in renal cancer (32). To the opposite, the knockout of CTNNB1 inhibits cell proliferation through the Wnt/β-catenin signaling pathway (33). Since the vital role of CTNNB1 mutation in the progression of cancer development, the prognostic value of CTNNB1 has been accepted in cancers such as lung cancer (23). Noriaki Sungaga et al. showed that CTNNB1 mutation mediated constitutive activation of the Wnt signaling pathway was implicated in the progression of lung cancer, preferentially in LUAD (22). In this study, the cytological effects CTNNB1 S37C mutation in LUAD cells were investigated. We found that CTNNB1 S37C mutation contributed the proliferation and migratory properties of H1299 and H1975 cells. Furthermore, we performed transcriptome sequencing to investigate the potential molecular mechanism, and a total of 1,060 DEGs were screened between NC and CTNNB1 S37C mutation groups, such as PTPRD, PTPRU, IGFBP3, MMP1 and CTNN1, etc. These DEGs were mainly enriched in functions such as extracellular matrix, as well as pathways including nicotine addiction.
Changes in gene expression is closed related with the development of LUAD (34). IGFBP-3, a protein interacted with cell-surface proteins, affects cell signaling from outside the cell or after internalization (35). It has been proved that IGFBP-3 can modulate the tumorigenesis and cell growth via IGF1 signaling in LUAD (36). Also, IGFBP-3 could interact with the extracellular matrix proteins (37). A previous study shows that the basal methylation status of IGFBP-3 could serve as a biomarker in clinical and could be used to predict the chemotherapy outcome in lung cancer (38). MMP1 is a extracellular matrix protein which vital for cell behaviors such as cell proliferation, migration (adhesion/dispersion), differentiation, angiogenesis, apoptosis, and host defense (39). The up-regulation of MMP-1 contribute to the malignant tumor progression in LUAD (40). Satio et al. showed that MMP-1 was an important therapeutic target for drug development in LUAD (41).
Actually, the biological function of genetic mutation or differentially expressed gene in tumor cell are commonly realized via certain functions and pathways (42,43). Extracellular matrix function act a crucial role in the progression of lung cancer (44). It has been proved that immunohistochemical expression of extracellular matrix proteins is correlated with lymph node metastasis in lung cancer (45). A previous study shows that the alterations in extracellular matrix function lead to the development of both metastasizing and non-metastasizing LUAD (46). In this study, PPI network showed that IGFBP-3 interacted with MMP-1. Therefore, we speculated IGFBP-3 and MMP-1 might play roles in the effect of CTNNB1 S37C mutation in LUAD by regulating extracellular matrix.
PTPRD and PTPRU are members of protein tyrosine phosphatase family that play roles in the regulation of cell proliferation and differentiation, oncogenic transformation and so on (47). PTPRU has been reported to serve as a regulator to negatively regulate the adhesion and proliferation in certain cancer cell types, mainly by its dephosphorylation of β-catenin and inhibition of following downstream signals (48). A previous study indicates that endogenous PTPRU promotes glioma progression through their effect on β-catenin and focal adhesion signaling (49). Via targeting PTPRU, certain miRNA is proved to promote metastasis in lung cancer (50). We concluded that PTPRD and PTPRU might play roles in the effect of CTNNB1 S37C mutation in LUAD. However, all the results were obtained based on data analysis. Basic experiments are needed to confirm these findings. In addition, there still remain some limitations in current study. For examples, only CTNNB1 S37C mutation was investigated. Also, we were unable to explore a large sample and lack of results of clinical samples. Thus, a further verification analysis based on a large sample size is needed.
In conclusion, CTNNB1 S37C mutation contributed the LUAD cells proliferation and migration. PTPRD, IGFBP-3, MMP1 and PTPRU might play roles in the effect of CTNNB1 S37C mutation in LUAD. Our study demonstrated the cytological effects of CTNNB1 S37C mutation in LUAD, and provided potential targets to investigate the molecular mechanisms.
Acknowledgments
Funding: The work was supported by grants from the Shanghai Science and Technology Commission (no. 20035800100).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-1146
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-1146
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-1146). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Senosain MF, Massion PP. Intratumor Heterogeneity in Early Lung Adenocarcinoma. Front Oncol 2020;10:349. [Crossref] [PubMed]
- Zhang L, Chen J, Cheng T, et al. Identification of the key genes and characterizations of Tumor Immune Microenvironment in Lung Adenocarcinoma (LUAD) and Lung Squamous Cell Carcinoma (LUSC). J Cancer 2020;11:4965-79. [Crossref] [PubMed]
- Yang D, He Y, Wu B, et al. Predictions of the dysregulated competing endogenous RNA signature involved in the progression of human lung adenocarcinoma. Cancer Biomark 2020;29:399-416. [Crossref] [PubMed]
- Yang W, You N, Jia M, et al. Undetectable circulating tumor DNA levels correlate with low risk of recurrence/metastasis in postoperative pathologic stage I lung adenocarcinoma patients. Lung Cancer 2020;146:327-34. [Crossref] [PubMed]
- Gong L, Hu Y, He D, et al. Ubiquitin ligase CHAF1B induces cisplatin resistance in lung adenocarcinoma by promoting NCOR2 degradation. Cancer Cell Int 2020;20:194. [Crossref] [PubMed]
- Han F, Huang D, Huang X, et al. Exosomal microRNA-26b-5p down-regulates ATF2 to enhance radiosensitivity of lung adenocarcinoma cells. J Cell Mol Med 2020;24:7730-42. [Crossref] [PubMed]
- Hammouz RY, Kostanek JK, Dudzisz A, et al. Differential expression of lung adenocarcinoma transcriptome with signature of tobacco exposure. J Appl Genet 2020;61:421-37. [Crossref] [PubMed]
- Tessema M, Tassew DD, Yingling CM, et al. Identification of novel epigenetic abnormalities as sputum biomarkers for lung cancer risk among smokers and COPD patients. Lung Cancer 2020;146:189-96. [Crossref] [PubMed]
- Leduc N, Ahomadegbe C, Agossou M, et al. Incidence of lung adenocarcinoma biomarker in a Caribbean and African Caribbean population. Journal of Thoracic Oncology 2016;11:769-73. [Crossref] [PubMed]
- D'Angelo SP, Pietanza MC, Johnson ML, et al. Incidence of EGFR exon 19 deletions and L858R in tumor specimens from men and cigarette smokers with lung adenocarcinomas. J Clin Oncol 2011;29:2066-70. [Crossref] [PubMed]
- Costigan DC, Dong F, Nucci MR, et al. Clinicopathologic and immunohistochemical correlates of CTNNB1 mutated endometrial endometrioid carcinoma. Int J Gynecol Pathol 2020;39:119-27. [Crossref] [PubMed]
- Akyol A, Güner G, Özşeker HS, et al. An immunohistochemical approach to detect oncogenic CTNNB1 mutations in primary neoplastic tissues. Lab Invest 2019;99:128-37. [Crossref] [PubMed]
- Colombo C, Miceli R, Lazar AJ, et al. CTNNB1 45F mutation is a molecular prognosticator of increased postoperative primary desmoid tumor recurrence: an independent, multicenter validation study. Cancer 2013;119:3696-702. [Crossref] [PubMed]
- Yao H, Ashihara E, Maekawa T. Targeting the Wnt/β-catenin signaling pathway in human cancers. Expert Opin Ther Targets 2011;15:873-87. [Crossref] [PubMed]
- Kim W, Kim M, Jho E-h. Wnt/β-catenin signalling: from plasma membrane to nucleus. Biochem J 2013;450:9-21. [Crossref] [PubMed]
- Zhang N, Wei P, Gong A, et al. FoxM1 promotes β-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell 2011;20:427-42. [Crossref] [PubMed]
- Yu W, Ma Y, Shankar S, et al. SATB2/β-catenin/TCF-LEF pathway induces cellular transformation by generating cancer stem cells in colorectal cancer. Sci Rep 2017;7:10939. [Crossref] [PubMed]
- Tang X, Zheng D, Hu P, et al. Glycogen synthase kinase 3 beta inhibits microRNA-183-96-182 cluster via the β-Catenin/TCF/LEF-1 pathway in gastric cancer cells. Nucleic Acids Res 2014;42:2988-98. [Crossref] [PubMed]
- Mencej-Bedrač S, Preželj J, Kocjan T, et al. Analysis of association of LRP5, LRP6, SOST, DKK1, and CTNNB1 genes with bone mineral density in a Slovenian population. Calcif Tissue Int 2009;85:501-6. [Crossref] [PubMed]
- Kono M, Bandoh N, Matsuoka R, et al. Glomangiopericytoma of the nasal cavity with CTNNB1 p. S37C mutation: a case report and literature review. Head Neck Pathol 2019;13:298-303. [Crossref] [PubMed]
- Braggio D, Zewdu A, Londhe P, et al. beta-catenin S45F mutation results in apoptotic resistance. Oncogene 2020;39:5589-600. [Crossref] [PubMed]
- Sunaga N, Kohno T, Kolligs FT, et al. Constitutive activation of the Wnt signaling pathway by CTNNB1 (β‐catenin) mutations in a subset of human lung adenocarcinoma. Genes Chromosomes Cancer 2001;30:316-21. [Crossref] [PubMed]
- Woenckhaus M, Merk J, Stoehr R, et al. Prognostic value of FHIT, CTNNB1, and MUC1 expression in non-–small cell lung cancer. Hum Pathol 2008;39:126-36. [Crossref] [PubMed]
- Zhou C, Li W, Shao J, et al. Analysis of the clinicopathologic characteristics of lung adenocarcinoma with CTNNB1 mutation. Front Genet 2020;10:1367. [Crossref] [PubMed]
- Livak KJ ST. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Kurnit K, Fellman B, Xie S, et al. The impact of adjuvant treatment in intermediate risk, stage I endometrial cancer with somatic CTNNB1 mutation. Gynecol Oncol 2019;153:e8 [Crossref]
- Imboden S, Tapia C, Scheiwiller N, et al. Early‐stage endometrial cancer, CTNNB1 mutations, and the relation between lymphovascular space invasion and recurrence. Acta Obstet Gynecol Scand 2020;99:196-203. [Crossref] [PubMed]
- Liu X, Wang L, Zhao S, et al. β-Catenin overexpression in malignant glioma and its role in proliferation and apoptosis in glioblastma cells. Med Oncol 2011;28:608-14. [Crossref] [PubMed]
- Wang G, Cui T, Sun L, et al. Long noncoding RNA LeXis promotes osteosarcoma growth through upregulation of CTNNB1 expression. Am J Cancer Res 2017;7:1577-87. [PubMed]
- Yang CM, Ji S, Li Y, et al. β-Catenin promotes cell proliferation, migration, and invasion but induces apoptosis in renal cell carcinoma. Onco Targets Ther 2017;10:711. [Crossref] [PubMed]
- Guan L, Zhu S, Han Y, et al. Knockout of CTNNB1 by CRISPR-Cas9 technology inhibits cell proliferation through the Wnt/β-catenin signaling pathway. Biotechnol Lett 2018;40:501-8. [Crossref] [PubMed]
- Hua X, Zhao W, Pesatori AC, et al. Genetic and epigenetic intratumor heterogeneity impacts prognosis of lung adenocarcinoma. Nat Commun 2020;11:2459. [Crossref] [PubMed]
- Yang G, Zhao W, Qin C, et al. Igfbp3 in grass carp (Ctenopharyngodon idellus): Molecular identification and mRNA expression under glucose, insulin and glucagon. Comp Biochem Physiol B Biochem Mol Biol 2020;242:110394 [Crossref] [PubMed]
- Wang YA, Sun Y, Palmer J, et al. IGFBP3 modulates lung tumorigenesis and cell growth through IGF1 signaling. Mol Cancer Res 2017;15:896-904. [Crossref] [PubMed]
- Varma Shrivastav S, Bhardwaj A, Pathak KA, et al. Insulin-Like Growth Factor Binding Protein-3 (IGFBP-3): Unraveling the Role in Mediating IGF-Independent Effects Within the Cell. Front Cell Dev Biol 2020;8:286. [Crossref] [PubMed]
- Ibanez de Caceres I, Cortes-Sempere M, Moratilla C, et al. IGFBP-3 hypermethylation-derived deficiency mediates cisplatin resistance in non-small-cell lung cancer. Oncogene 2010;29:1681-90. [Crossref] [PubMed]
- Wang LE, Huang YJ, Yin M, et al. Promoter polymorphisms in matrix metallopeptidase 1 and risk of cutaneous melanoma. Eur J Cancer 2011;47:107-15. [Crossref] [PubMed]
- Schütz A, Röser K, Klitzsch J, et al. Lung adenocarcinomas and lung cancer cell lines show association of MMP-1 expression with STAT3 activation. Transl Oncol 2015;8:97-105. [Crossref] [PubMed]
- Saito R, Miki Y, Ishida N, et al. The Significance of MMP-1 in EGFR-TKI–Resistant Lung Adenocarcinoma: Potential for Therapeutic Targeting. Int J Mol Sci 2018;19:609. [Crossref] [PubMed]
- Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer 2006;118:257-62. [Crossref] [PubMed]
- Li W, Zhang M, Huang C, et al. Genetic variants of DNA repair pathway genes on lung cancer risk. Pathol Res Pract 2019;215:152548 [Crossref] [PubMed]
- Götte M, Kovalszky I. Extracellular matrix functions in lung cancer. Matrix Biology 2018;73:105-21. [Crossref] [PubMed]
- Han J-Y, Kim HS, Lee SH, et al. Immunohistochemical expression of integrins and extracellular matrix proteins in non-small cell lung cancer: correlation with lymph node metastasis. Lung Cancer 2003;41:65-70. [Crossref] [PubMed]
- Clarke MR, Landreneau RJ, Finkelstein SD, et al. Extracellular matrix expression in metastasizing and nonmetastasizing adenocarcinomas of the lung. Hum Pathol 1997;28:54-9. [Crossref] [PubMed]
- Kim M, Morales LD, Jang IS. Protein Tyrosine Phosphatases as Potential Regulators of STAT3 Signaling. Int J Mol Sci 2018;19:2708. [Crossref] [PubMed]
- Gu J, Zhang Z, Lang T, et al. PTPRU, As A Tumor Suppressor, Inhibits Cancer Stemness By Attenuating Hippo/YAP Signaling Pathway. Onco Targets Ther 2019;12:8095-104. [Crossref] [PubMed]
- Zhu Z, Liu Y, Li K, et al. Protein tyrosine phosphatase receptor U (PTPRU) is required for glioma growth and motility. Carcinogenesis 2014;35:1901-10. [Crossref] [PubMed]
- Zhou R, Zhou X, Yin Z, et al. MicroRNA-574-5p promotes metastasis of non-small cell lung cancer by targeting PTPRU. Sci Rep 2016;6:35714. [Crossref] [PubMed]
(English Language Editor: D. Fitzgerald)


