
Co-culturing polarized M2 Thp-1-derived macrophages enhance stemness of lung adenocarcinoma A549 cells
Introduction
Lung cancer is the predominant cause of cancer-related mortality worldwide. Non-small cell lung cancer (NSCLC) is the most common type of lung cancer, making up about 80% of all lung cancer cases. At present, the treatment of NSCLC mainly involves surgical resection, radiotherapy, chemotherapy, biological immunotherapy, and molecular targeted therapy. Of these, surgical resection is still the most effective method for the treatment of NSCLC. However, almost 70–80% of patients are diagnosed at a late stage, and only 20–30% of patients have indications of operation. Even worse, the postoperative recurrence and metastasis rates are still as high as 50%. The majority of patients with advanced NSCLC have poor sensitivity to radiotherapy. Recently, molecular targeted therapy has become a critical focus of tumor research due to its high efficiency and low adverse reactions. Epidermal growth factor receptor (EGFR)-targeted therapy for NSCLC is increasingly prominent and has become a hotspot for global research (1,2). Targeted drugs represented by EGFR tyrosine kinase inhibitors (TKI), such as Gefitinib, Osimertinib, and Afatinib, have significantly extended the survival time of advanced NSCLC patients, however acquired resistance inevitably follows short-term effective therapy (1,2). Therefore, other molecular targets for gene therapy of advanced NSCLC patients are urgently needed.
Previous studies have demonstrated that the tumor microenvironment (TME) is vital for the occurrence, growth, and metastasis of tumors (3). It is complex and consists of multiple cell types containing tumor-associated macrophages (TAMs) (4). An increasing number of studies have revealed that TAMs could increase the tumorigenesis, metastasis, and invasion of cancer cells, including lung cancers (5-8). A study by Che et al. reported that when co-cultured with A549 cells, macrophages could induce epithelial-mesenchymal transition (EMT) and invasion of A549 cells via the PGE2/ COX-2/IL-6/β-catenin signaling pathway (7). These findings highlight the significance of exploring the function and biological behavior of TAMs in cancer.
As a subpopulation of tumor cells, cancer stem cells (CSCs) are related to tumor progression, metastasis, resistance, and recurrence, primarily through key signaling pathways such as the Hedgehog, Notch, Wnt, and Signal Transducer and Activator of Transcription 3 (STAT3) pathways (9-11). Recent studies have revealed that lung cancer stem cells (LCSCs) play critical roles in tumor progression, metastasis, and drug resistance in lung cancer (12-14). A study by Sun et al. showed that sphere-derived NSCLC cells with enhanced expression of the CSC marker, CD133, exhibited increased proliferation, cell-cycle progression, and drug-resistance (12). Moreover, Park et al. found that the expression of the CSC marker, NANOG, was correlated with high histological grade, lymphatic invasion, EMT of lung adenocarcinoma, and was an independent risk factor for poor prognosis in lung adenocarcinoma (14). Therefore, investigating LCSCs will help us understand the mechanisms and discover more potential molecular targets for the treatment of lung cancer.
Recent studies have demonstrated that macrophage-conditioned medium could promote the stemness of several cancers, including colorectal cancer, ovarian cancer, renal cell carcinoma, and breast cancer via the STAT3, hedgehog, and AKT/mTOR signaling pathways (15-18). It has been reported that the expression of the cancer stemness gene CD133 and the self-renewal ability of the NSCLC cells were significantly enhanced in the presence of M2-tumor-associated macrophages (M2-TAMs), suggesting that M2-TAMs could promote the generation of CSCs (19). The interaction between macrophages and CSCs may play a vital role in the development and prognosis of lung cancer. However, up to now, the underlying molecular mechanism through which TAMs interact with the stemness of lung cancer is unclear.
In this study, the viability and stemness of A549 cells and the activation of macrophages were examined in a co-culture system by using the Hedgehog inhibitor, Vismodegib, the Notch inhibitor, Gamma Secretase Inhibitor (GSI), and the STAT3 inhibitor, Cucurbitacin I (JSI-124). Our study demonstrated that the co-culture system could induce stemness of A549 cells via the STAT3, Notch, and Hedgehog pathways. It also identifies a new role of TAMs in the regulation of LCSCs and suggests potential therapeutic strategies for lung cancer therapy in the future. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-21-1256).
Methods
Cell culture
The monocyte cell lines, THP-1, and the human NSCLC cell lines, A549, were obtained from Cell Bank of Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Roswell park memorial institute (RPMI) 1640 medium (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS, Gibco, Detroit, MI, USA), penicillin (100 U/mL; Invitrogen, USA), and streptomycin (100 µg/mL; Invitrogen, USA). The culture environment was maintained at 37 °C in a humidified 5% carbon dioxide (CO2) atmosphere. The subcultured monocyte THP-1 cells were seeded at 1×106/mL and induced with 100-ng/mL phorbol-12-myristate-13-acetate (PMA, Sigma Chemical, St. Louis, MO, USA) for 72 h to adhere and generate THP-1-derived macrophages.
Co-culture procedures
The co-culture system was performed using a cell culture insert (0.4-µm porous; Corning, NY, USA). First, the THP-1 monocytes (1×106 cells/mL) were seeded into the upper chamber of the transwell device and treated with 100 ng/mL PMA for 72 h to stimulate differentiation. Next, the cells were washed with phosphate-buffered saline (PBS) three times and cultured for 24 h to exclude the interference of PMA. The A549 cells (1×105 cells/mL) were seeded in the lower chamber and incubated for 24 h to allow for attachment. Subsequently, the chambers containing the THP-1-derived macrophages were directly placed on top of the six-well plates with the A549 cells, and the resulting co-culture system cells were cultured with RPMI-1640 medium for 24 h. The A549 cells and THP-1-derived macrophages were separately incubated with RPMI-1640 medium in six-well plates for 24 h as the corresponding controls.
Cell Counting Kit-8 (CCK-8) assay
The effect of co-culturing with THP-1-derived macrophages on the viability of A549 cells was examined using a CCK-8 assay kit (Dojindo Laboratories, Kumamoto, Japan). In this experiment, the A549 cells were incubated with various percentage concentrations of THP-1-derived macrophages (100:1, 10:1, 1:1) in RPMI 1640 medium. The effect of STAT3, Notch, and Hedgehog inhibitors on the viability of the co-culture system was also investigated. Co-culture system cells were incubated with various concentrations of Hedgehog inhibitor (Vismodegib) (0.1, 1, 10 µM), Notch inhibitor (GSI) (2, 5, 10 µM), and STAT3 inhibitor (JSI-124) (1, 5, 10 µM). A549 cells of each group were digested and seeded in 96-well plates (1,000 cells/well, n=6). After 48 h, 10 µL CCK8 solution was added to each well and cultured for another 4 h at 37 °C. Finally, the absorbance at 450nm was determined using a microplate reader (Bio-Rad, Hercules, CA, USA), and the proliferation was calculated using the following equation: proliferation rate (%) = A (sample)/A (control) × 100.
Cell-IQ proliferation and tracking
The cell growth and morphological changes were recorded and analyzed using a Cell-IQ cell culturing platform (Chip-Man Technologies, Tampere, Finland), equipped with a phase-contrast microscope (Nikon CFI Achromat phase-contrast objective with 10 magnification) and a camera (20). The THP-1-derived macrophages, A549 cells, co-culture system, co-culture system with 10µM Vismodegib, co-culture system with 5 µM GSI, and co-culture system with 10 µM JSI-124 were inoculated into 24-well plates at a density of 30,000 cells/well and then maintained at 37 °C in a humidified 5% CO2 Cell-IQ incubator. Digital photographs of the cell cultures were obtained every 45 min, and every 16th image was used for an automated analysis with the Cell-IQ Image software. Cell-IQ analysis software was then used to analyze the cell growth and morphological changes. Finally, the growth was evaluated using the following equation: increased fold of total cell number = [T(n) − T(0)]/T(0), where T(n) is the total cell number at n time point, and T(0) is the initial total cell number.
RNA isolation and qRT-PCR analysis
Total ribonucleic acid (RNA) from the cultured A549 cells, THP-1-derived macrophages, co-culture system, co-culture system with 10 µM Vismodegib, co-culture system with 5 µM GSI, and co-culture system with 10 µM JSI-124 were extracted using Trizol reagent (Invitrogen, USA) according to the manufacturer’s instructions. Complementary deoxyribonucleic acids (cDNAs) were then synthetized using the PrimeScript RT reagent kit (TaKaRa, Dalian, China) and quantified using the SYBR Premix Ex Taq (TaKaRa, Dalian, China) according to the manufacturer’s instructions. Quantitative real-time polymerase chain reaction (qRT-PCR) reactions were carried out using StepOne Plus™ Real-Time PCR System (Thermo Fisher Scientific, Waltham, USA). The relative expression was calculated using the 2−ΔΔCt method, where Ct represents the threshold cycle. GAPDH was used as the internal control.
Flow cytometry analysis
The activity of the Acetaldehyde dehydrogenase (ALDH) enzyme was detected using an Aldefluor Assay Kit (Stem Cell Technologies, Canada) according to the manufacturer’s instructions. Briefly, the cells were suspended in Aldefluor assay buffer and incubated with ALDH enzyme substrate bodipyaminoacetaldehyde (BAAA) for 50 min at 37 °C. For the control, cells pre-treated with the ALDH-specific inhibitor, diethylaminobenzaldehyde (DEAB), were resuspended in Aldefluor assay buffer and incubated with BAAA. Finally, the cells were analyzed on a FACSCalibur flow cytometry (BD Biosciences, San Jose, CA, USA).
Enzyme-linked immunosorbent assay (ELISA)
The supernatants from the A549 cells, THP-1-derived macrophages, co-culture system, co-culture system with 10µM Vismodegib, co-culture system with 5 µM GSI and co-culture system with 10µM JSI-124 were collected in RPMI 1640 medium after incubation for 24 h. Next, the medium was removed the cellular debris by centrifugation, and was reserved at −80ºC until it was subjected to ELISA to examine the protein expression levels of Transforming growth factor-β (TGF-β) and Interleukin-6 (IL-6). The concentrations of TGF-β and IL-6 were detected using respective ELISA kits (Boster Biological Technology, Pleasanton, CA, USA) according to the manufacturer’s instructions.
Western blot analysis
Cells were collected and lysed using RIPA Lysis Buffer (Beyotime, China), and added with protease inhibitor cocktail (Roche, Basel, Switzerland) and phenylmethylsulfonyl fluoride (PMSF, Sigma Chemical). The lysates were centrifugated at 12,000 rpm for 5 min and the supernatants were collected. Subsequently, protein content was measured using a Bradford assay. 50 µg protein extractions were separated by 12% SDS-PAGE (Beyotime), and were then transferred to Polyvinylidene fluoride (PVDF) Immobilon-P-membrane (Millipore, Burlington, MA, USA) and incubated with the respective antibodies. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody was used as an internal control. The bands were visualized by enhanced chemiluminescent detection kit (Millipore, USA) and subsequently exposed using the Molecular Imager ChemiDoc XRS System (Bio-Rad, USA). The antibodies of SOX2, NANOG, OCT4, CD44, TGF-β and GAPDH as well as the secondary antibody were obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA).
Statistical analyses
All data were analyzed using SPSS 17.0 (SPSS, Chicago, IL, USA) and GraphPad Prism 8.0 (GraphPad Software, La Jolla, CA, USA). Data from the quantitative assays were presented as means ± standard deviation and each experiment was conducted in triplicate. Significant differences between the groups were analyzed using one-way analysis of variance (ANOVA), and the Duncan test was used for multiple comparisons. P<0.05 was considered to be statistically significant and asterisks were used to indicate different levels of significance (*P<0.05, **P<0.01).
Results
Effect of co-culturing with THP-1-derived macrophages on the viability of A549 cells
The effect of co-culturing with THP-1-derived macrophages on the viability of A549 cells was examined using CCK-8 assay. In this experiment, the A549 cells were cultured with various percentage concentrations of THP-1-derived macrophages (100:1, 10:1, 1:1) in RPMI 1640 medium. The results showed that the proliferation of A549 cells was obviously enhanced after co-culturing with THP-1-derived macrophages. The proliferation rates of A549 cells at the aforementioned percentage concentrations (100:1, 10:1 and 1:1) were 1.36%, 47.11%, and 56.32%, respectively. There was a statistically significant difference between the A549 cells and the co-culture system at the 10:1 and 1:1 percentage concentration (Figure 1A, P<0.01). Therefore, the co-culture system (10:1) was selected for subsequent Cell-IQ, qRT-PCR, flow cytometry, western blotting, and ELISA assay analyses.
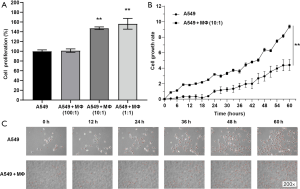
The Cell-IQ continuous surveillance system was used to observe cell growth and morphological changes. This system for continuous monitoring of living cells is a promising tool in cellular biology as it allows researchers to study morphology in vitro and explore behavioral responses of cells in real time. As shown in Figure 1B, the proliferation of A549 cells was significantly enhanced after co-culturing with THP-1-derived macrophages. The red dots in the figure means stable cells in the Cell-IQ system and count the number of stable cells (Figure 1C). We counted dead cells under electron microscope by cell morphology. The number of dead cells was obviously reduced after co-culturing with THP-1-derived macrophages.
Co-culturing with THP-1-derived macrophages induced stemness of A549 cells
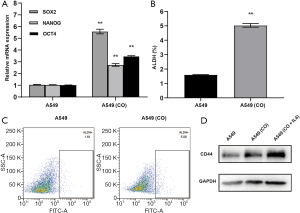
To examine the effect of THP-1-derived macrophages on the stemness of A549 cells, qRT-PCR assays were used to analyze the messenger ribonucleic acid (mRNA) levels of CSC markers SOX2, NANOG, and OCT4 of A549 cells. Compared with the A549 cells, the co-culture system significantly increased the mRNA level of SOX2 by 5.38 folds, NANOG by 2.66 folds, and OCT4 by 3.42 folds (Figure 2A, P<0.01). As a CSC marker, ALDH activity was investigated by flow cytometry analysis. Cells with a high expression of ALDH were marked with green fluorescent protein. Compared with the A549 cells, the proportion of ALDH positive cells was markedly increased in the co-culture system (from 1.61% to 5.09%) (Figure 2B,C, P<0.01). We treated co-culture systems with IL-4 to be a positive control group (M2 phenotype) comparing with co-culture treatment alone group. Western blot results indicated that expression of CD44 proteins was obviously increased in the co-culture systems and co-culture systems treated with IL-4 compared with the A549 cells (Figure 2D).
Macrophage activation and polarization after co-culturing with A549 cells
To verify whether the co-culture system affected macrophage activation, qRT-PCR and ELISA assays were performed to measure the mRNA levels of TGF-β and IL-6. Compared with the macrophages, the co-culture system notably enhanced the mRNA expression levels of TGF-β by 4.48 folds and IL-6 by 4.81 folds (Figure 3A, P<0.01). Moreover, the protein expression levels of TGF-β and IL-6 in macrophages were 6.31±0.83 and 2.61±0.38 ng/mL, respectively. The expression levels of TGF-β and IL-6 in the co-culture system were 22.19±0.97 and 11.84±0.77 ng/mL, respectively. Meanwhile, the levels of TGF-β and IL-6 in the supernatants were also significantly enhanced after co-culturing (Figure 3B, P<0.01).
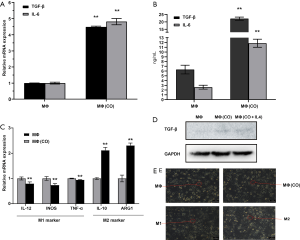
To explore effect of A549 cells on the differentiation of the macrophages, qRT-PCR assays were used to analyze the mRNA levels of M1-macrophage markers IL-12, TNF-α, INOS, and M2-macrophage markers IL-10 and ARG1. Compared with THP-1-derived macrophages, the co-culture of macrophages significantly decreased the mRNA expression level of IL-12 by 20.57%, INOS by 26.27%, and TNF-α by 5.66% (Figure 3C, P<0.01). In addition, the co-culture system significantly increased the mRNA level of IL-10 by 2.12 folds and ARG1 by 2.31 folds (P<0.01). We treated co-culture systems with IL-4 to be a positive control group (M2 phenotype) comparing with co-culture treatment alone group. Western blot results indicated that expression of TGF-β proteins was obviously increased in the co-culture systems and co-culture systems treated with IL-4 compared with the macrophages (Figure 3D). M1 and M2 macrophages induced by LPS/IFN-γ and IL-4. The M1 macrophages exhibit more oval while M2 macrophage demonstrate elongated spindle-like morphologies. The co-cultured macrophages display a morphology of elongated fusiform similar to M2 macrophages (Figure 3E).
Effect of inhibition of stemness-related signaling pathways on the growth and morphological changes of the co-culture system
To explore the molecular mechanisms between macrophage activation and the stemness of A549 cells, the effects of STAT3, Notch, and Hedgehog inhibitors on the viability of the co-culture system were examined. Co-culture system cells were incubated with different concentrations of Hedgehog inhibitor (Vismodegib) (0.1, 1, 10 µM), Notch inhibitor (GSI) (2, 5, 10 µM), and STAT3 inhibitor (JSI-124) (1, 5, 10 µM) in RPMI 1640 medium. The results of the CCK-8 assay indicated that the proliferation of the co-culture system was inhibited by the STAT3, Notch, and Hedgehog inhibitors. After 48 h, the inhibitory rates of Vismodegib at doses of 0.1, 1, and 10 µM were 7.26%, 10.72%, and 31.94%, respectively. There was statistical significance between the co-culture system and the co-culture system with 0.1, 1, and 10 µM Vismodegib (Figure 4A, P<0.01). The inhibitory rates of GSI at doses of 5 µM and 10 µM were 26.09% and 29.59%, respectively, which was statistically significant (Figure 4B, P<0.01). No inhibitory effect of GSI was observed at a dose of 2µM. The inhibitory rates of JSI-124 at doses of 1, 5 and 10 µM were 15.84%, 25.16%, and 34.02%, respectively, which was statistically significant (Figure 4C, P<0.01). Taken together, Vismodegib at dose of 10 µM, GSI at dose of 5 µM, and JSI-124 at dose of 10 µM were selected for subsequent Cell-IQ, qRT-PCR, flow cytometry, western blotting, and ELISA assay analyses.
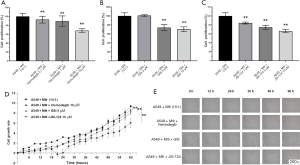
The results of Cell-IQ proliferation and tracking showed that the proliferation of the co-culture system was significantly reduced after inhibition of stemness-related signaling pathways. The growth inhibitory effect of Vismodegib increased after 24 h, while the growth inhibitory effect of GSI almost disappeared after 36 h (Figure 4D). Furthermore, the growth inhibitory effect of JSI-124 increased in a time-dependent manner (Figure 4D). The number of dead cells was markedly increased after inhibition of stemness-related signaling pathways (Figure 4E).
Effect of inhibition of stemness-related signaling pathways on the stemness of A549 cells
In this experiment, qRT-PCR, flow cytometry analysis, and western blotting were performed to detect the stemness of A549 cells upon inhibition of the Hedgehog, Notch, and STAT3 signaling pathways. Compared with the co-culture system, the mRNA expression of SOX2, NANOG, and OCT4 were significantly reduced in the inhibitor treatment co-culture systems (Figure 5A, P<0.01). The co-culture system with 10 µM Vismodegib significantly decreased the mRNA expression level of SOX2 by 12.03%, NANOG by 50.96%, and OCT4 by 49.53% (P<0.01). The co-culture system with 5µM GSI notably reduced the mRNA expression level of SOX2 by 18.56%, NANOG by 56.98%, and OCT4 by 55.32% (P<0.01). The co-culture system with 10 µM JSI-124 markedly downregulated the mRNA expression level of SOX2 by 60.67%, NANOG by 22.17%, and OCT4 by 28.77% (P<0.01). As shown above, the STAT3 inhibitor exerted the strongest inhibitory effect on SOX2 expression (Figure 5A, P<0.01), while the Notch inhibitor exhibited the strongest inhibitory effect on the expression of NANOG and OCT4 (Figure 5A, P<0.01). Compared with the co-culture system, the proportion of ALDH positive cells was significantly decreased in the inhibitor treatment co-culture systems (Figure 5B,C, P<0.01). Western blot results indicated that expressions of SOX2, NANOG, and OCT4 proteins were obviously downregulated in the inhibitor treatment co-culture systems compared with the co-culture systems (Figure 5D).
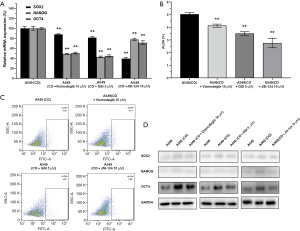
Evaluation of macrophage activation following inhibition of stemness-related signaling pathways
QRT-PCR and ELISA assays were used to assess the levels of TGF-β and IL-6 in the co-culture systems. The co-culture system with 10 µM Vismodegib significantly decreased the mRNA expression level of TGF-β by 37.04%, IL-6 by 8.43% (Figure 6A, P<0.01). The co-culture system with 5 µM GSI notably reduced the mRNA expression level of TGF-β by 56.06%, IL-6 by 38.87% (Figure 6A, P<0.01). The co-culture system with 10 µM JSI-124 significantly downregulated the mRNA expression level of TGF-β by 66.10% and IL-6 by 64.89% (Figure 6A, P<0.01). Compared with the co-culture system, the mRNA expression levels of TGF-β and IL-6 were notably downregulated in the inhibitor treatment co-culture systems (Figure 6A, P<0.01).

The expression level of TGF-β in the co-culture system with 10 µM Vismodegib, co-culture system with 5 µM GSI, and co-culture system with 10µM JSI-124 were 11.59±1.99, 13.89±0.81 and 8.55±1.04 ng/mL, respectively, which were significantly lower than those of co-culture system (Figure 6B, P<0.01). The expression level of IL-6 in the co-culture system with 10 µM Vismodegib, co-culture system with 5 µM GSI, and co-culture system with 10 µM JSI-124 were 11±1.22, 11.63±0.82 and 4.32±0.63 ng/mL, respectively. Compared with the co-culture system, only the co-culture system with 10 µM JSI-124 suppressed the IL-6 expression level significantly (Figure 6B, P<0.01). qRT-PCR was used to assess the mRNA levels of IL-6. There is a complicated process from mRNA to protein, so the expression of mRNA and protein are not necessarily positively correlated. As shown above, the STAT3 inhibitor had the strongest inhibitory effect on the expression of the tumor-promoting genes, TGF-β and IL-6.
Discussion
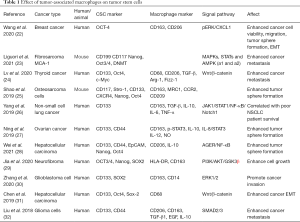
Previous studies have demonstrated that M2-TAMs could enhance the proliferation, angiogenesis, EMT, migration and invasion of multiple cancers, including lung cancer, Breast cancer, osteosarcoma cells, ovarian cancer and so on (5,7,8,21). Recently, an increasing number of studies have suggested that TAMs could enhance CSC phenotypes and affects viability, metastasis, tumor sphere formation and EMT of multiple cancers (Table 1). Yang et al. reported that TAMs could induce CSC-like phenotypes in breast cancer cells by upregulating the expression of SOX2, NANOG, OCT4, AbcG2, and Sca-1, and that the EGFR and STAT3 inhibitors could effectively block macrophage-induced upregulation of SOX-2 and CSC phenotypes in breast cancer cells (15). Fan et al. reported that macrophage-conditioned medium could promote colorectal CSC phenotypes via the Hedgehog signaling pathway (16). Yang et al. found that infiltrating macrophages could increase the EMT, invasion and stem cell-like populations of renal cell carcinoma via activation of AKT/mTOR signaling (17). Ning et al. showed that the co-culture of THP-1 derived macrophages with ovarian cancer stem-like cells could promote M2-TAMs polarization and stemness of ovarian cancer cells via IL8/STAT3 signaling, and that upregulation of STAT3 could rescue the effects of IL-8 on M2-TAMs polarization and stemness of ovarian cancer cells (18). Macrophage-induced CSC phenotypes have also been found in lung cancer. A study by Huang et al. revealed that NSCLC cells exhibited significantly increased expression of cancer stemness gene CD133 and enhanced self-renewal ability in the presence of M2-TAMs (19). Therefore, the interaction between macrophages and CSCs may play an important role in the development and gene therapy of lung cancer (33,34). However, until now, the underlying molecular mechanism through which TAMs interact with stemness of lung cancer cells is still unclear.
Full table
In the present study, the proliferation of A549 cells was significantly enhanced after co-culturing with THP-1-derived macrophages. The mRNA expression levels of CSC markers SOX2, NANOG, and OCT4 in A549 cells were notably promoted and the proportion of ALDH-positive cells was significantly increased after co-culturing with THP-1-derived macrophages. Meanwhile, the expressions of TGF-β and IL-6 in macrophages co-cultured with A549 were notably elevated compared with those from THP-1-derived macrophages. Based on these findings, we hypothesized that TAMs may enhance the stemness of A549 cells by secreting TGF-β and IL-6. In this study, the Hedgehog, Notch, and STAT3 inhibitors reduced the proliferation and stemness of A549 cells, and also decreased the expression levels of TGF-β and IL-6 in macrophages co-cultured with A549 cells. These findings suggested that TAMs might promote the stemness of A549 cells via the STAT3, Notch, and Hedgehog signaling pathways. Hedgehog, as one of the predominant mediators involved in the maintenance and self-renewal of CSCs, has been revealed in many cancers, including lung cancer (35). The activation of STAT3 pathway was closely associated with the occurrence, transformation, invasion and metastasis of various tumors (36). Also, the inhibition of Notch signaling could induce apoptosis and reduce the proliferation and in vivo growth of lung cancer cells (37). In this study, the Hedgehog, Notch, and STAT3 inhibitors reduced the proliferation and stemness of A549 cells. Yet, whether Hedgehog, Notch, and STAT3 inhibitors affect the EMT, migration, and invasion of A549 cells needs to be further explored.
Typically, TAMs exhibit two different polarization types, including M1-TAMs and M2-TAMs. M1-TAMs can secrete pro-inflammatory cytokines such as IL-12 and TNF-α to inhibit tumor growth, while M2-TAMs secrete anti-inflammatory cytokines such as IL-10 and TGF-β to promote the growth, angiogenesis, metastasis, and invasion of tumors (38). Previous reports have demonstrated that TGF-β could promote the differentiation of non-activated macrophages into M2-TAMs (39,40). Also, TGF-β secreted by macrophages could regulate the growth, immune-suppression, angiogenesis, EMT, invasion, and chemoresistance of cancers, including lung cancer (41-46). These studies suggested that TGF-β plays an important regulatory role in the differentiation and function of TAMs. In the present study, we found that the expression of TGF-β was first elevated in the co-culture system and then decreased significantly following administration of the STAT3, Notch, and Hedgehog inhibitors. These results indicated that TGF-β may induce M2-TAMs polarization in lung cancer through the STAT3, Notch, and Hedgehog pathways.
As an inflammatory cytokine, IL-6 was enrich in the TME and stimulated the development of many different types of cancers (47). Increasing evidence suggested that IL-6/STAT3 pathway exerts vital effects in the macrophage polarization and tumor progression (48-53). Fu et al. found that IL-6 could induce the differentiation of normal macrophages into M2-TAMs with upregulated IL-10 and TGF-β levels via activating STAT3 phosphorylation, resulting in the promotion of gastric cancer cell proliferation and migration (48). Yin et al. found that the IL-6/STAT3 pathway was activated in M2-TAMs and that the inhibition of the IL-6/STAT3 pathway could turn macrophages into M1-TAMs, leading to reduced viability, proliferation, invasion, migration, and drug resistance in hepatocellular carcinoma (49). The secretion of IL-6 from mesenchymal stem cells could enhance the progression of colorectal and lung cancers via activation of the JAK2/STAT3 pathway (50,51). Furthermore, the IL-6/STAT3 and IL-6/Notch pathways were also shown to be involved in the promotion of stem cell-like properties of breast and hepatocellular carcinomas (52,53). In the present study, the level of IL-6 was first elevated in the co-culture system and subsequently decreased significantly following administration of the STAT3, Notch, and Hedgehog inhibitors, suggesting that IL-6 may induce polarization of M2-TAMs and modulate the CSC phenotype via the STAT3, Notch, and Hedgehog pathways.
Clinical studies have revealed that the expressions of SOX2, NANOG, OCT4, and the activity of ALDH were significantly correlated with pathological grade, EMT, and lymph node status of adenocarcinoma, and were independent risk factors for poor prognosis in adenocarcinoma (14,54). The results of the current study showed that the STAT3, Notch, and Hedgehog inhibitors could effectively downregulate the expression of ALDH activity and stemness genes (SOX2, NANOG, OCT4). Previous studies have elucidated that the Hedgehog, STAT3, and Notch pathways could promote the stemness and initiation of lung cancer (20,50,55). In this study, the inhibitory rate of JSI-124 on the expression of SOX2 was the highest (60.67%), significantly higher than that of GSI (18.56%), and the inhibitory rate of Vismodegib was the lowest (12.03%). The same inhibition trend occurred in the activity of ALDH. Based on these findings, we hypothesized that STAT3 signaling may be the main pathway in regulating the expression of SOX2 and ALDH. The inhibitory rate of GSI on the expression of NANOG was the highest (56.98%), significantly higher than that of Vismodegib (50.96%), and the inhibitory rate of JSI-124 was the lowest (22.17%). The same inhibition trend occurred in the expression of OCT4. These results indicated that Notch signaling may be the primary pathway regulating the expression of NANOG and OCT4.
Taken together, the results of the present study suggested that THP-1-derived macrophages may promote the stemness of A549 cells via the IL-6/STAT3/SOX2 (ALDH), IL-6/Notch/OCT4 (NANOG) signaling pathways. Therefore, our report identifies the role of TAMs in the regulation of LCSCs and suggests potential therapeutic strategies for lung cancer therapy in the future. Further research needs to be carried out to validate the proposed hypothetical regulatory signaling pathways. Also, the interaction mechanism between macrophage polarization and stemness of lung cancer cells needs to be further explored in vivo.
Acknowledgments
Funding: This article was supported by the National Nature Science Foundation of China (81770074, 81570075, 81400035), Zhejiang Provincial Natural Science Foundation (LZ15H010001, LY18H010006), Major Program of the National Key Research and Development Program of China (2016YFC1304000), Key Laboratory of Interventional Pulmonology of Zhejiang Province (2019E10014), Zhejiang Provincial Key Research and Development Program (2020C03067).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-1256
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-1256
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-1256). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang B, Zuo Z, Li F, et al. Gefitinib versus Docetaxel in Treated Non-small-cell Lung Cancer: A Meta-analysis. Open Med (Wars) 2017;12:86-91. [Crossref] [PubMed]
- Bollinger MK, Agnew AS, Mascara GP. Osimertinib: A third-generation tyrosine kinase inhibitor for treatment of epidermal growth factor receptor-mutated non-small cell lung cancer with the acquired Thr790Met mutation. J Oncol Pharm Pract 2018;24:379-88. [Crossref] [PubMed]
- Hu C, Chen M, Jiang R, et al. Exosome-related tumor microenvironment. J Cancer 2018;9:3084-92. [Crossref] [PubMed]
- Sawa-Wejksza K, Kandefer-Szerszen M. Tumor-Associated Macrophages as Target for Antitumor Therapy. Arch Immunol Ther Exp (Warsz) 2018;66:97-111. [Crossref] [PubMed]
- Zhu F, Li X, Chen S, et al. Tumor-associated macrophage or chemokine ligand CCL17 positively regulates the tumorigenesis of hepatocellular carcinoma. Med Oncol 2016;33:17. [Crossref] [PubMed]
- Zhang S, Che D, Yang F, et al. Tumor-associated macrophages promote tumor metastasis via the TGF-beta/SOX9 axis in non-small cell lung cancer. Oncotarget 2017;8:99801-15. [Crossref] [PubMed]
- Che D, Zhang S, Jing Z, et al. Macrophages induce EMT to promote invasion of lung cancer cells through the IL-6-mediated COX-2/PGE2/beta-catenin signalling pathway. Mol Immunol 2017;90:197-210. [Crossref] [PubMed]
- She L, Qin Y, Wang J, et al. Tumor-associated macrophages derived CCL18 promotes metastasis in squamous cell carcinoma of the head and neck. Cancer Cell Int 2018;18:120. [Crossref] [PubMed]
- Lytle NK, Barber AG, Reya T. Stem cell fate in cancer growth, progression and therapy resistance. Nat Rev Cancer 2018;18:669-80. [Crossref] [PubMed]
- Takebe N, Harris PJ, Warren RQ, et al. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol 2011;8:97-106. [Crossref] [PubMed]
- Thiagarajan PS, Zheng Q, Bhagrath M, et al. STAT3 activation by leptin receptor is essential for TNBC stem cell maintenance. Endocr Relat Cancer 2017;24:415-26. [Crossref] [PubMed]
- Sun FF, Hu YH, Xiong LP, et al. Enhanced expression of stem cell markers and drug resistance in sphere-forming non-small cell lung cancer cells. Int J Clin Exp Pathol 2015;8:6287-300. [PubMed]
- Hashida S, Yamamoto H, Shien K, et al. Acquisition of cancer stem cell-like properties in non-small cell lung cancer with acquired resistance to afatinib. Cancer Sci 2015;106:1377-84. [Crossref] [PubMed]
- Park E, Park SY, Sun PL, et al. Prognostic significance of stem cell-related marker expression and its correlation with histologic subtypes in lung adenocarcinoma. Oncotarget 2016;7:42502-12. [Crossref] [PubMed]
- Yang J, Liao D, Chen C, et al. Tumor-associated macrophages regulate murine breast cancer stem cells through a novel paracrine EGFR/Stat3/Sox-2 signaling pathway. Stem Cells 2013;31:248-58. [Crossref] [PubMed]
- Fan F, Wang R, Boulbes DR, et al. Macrophage conditioned medium promotes colorectal cancer stem cell phenotype via the hedgehog signaling pathway. PLoS One 2018;13:e0190070 [Crossref] [PubMed]
- Yang Z, Xie H, He D, et al. Infiltrating macrophages increase RCC epithelial mesenchymal transition (EMT) and stem cell-like populations via AKT and mTOR signaling. Oncotarget 2016;7:44478-91. [Crossref] [PubMed]
- Ning Y, Cui Y, Li X, et al. Co-culture of ovarian cancer stem-like cells with macrophages induced SKOV3 cells stemness via IL-8/STAT3 signaling. Biomed Pharmacother 2018;103:262-71. [Crossref] [PubMed]
- Huang WC, Chan ML, Chen MJ, et al. Modulation of macrophage polarization and lung cancer cell stemness by MUC1 and development of a related small-molecule inhibitor pterostilbene. Oncotarget 2016;7:39363-75. [Crossref] [PubMed]
- Song D, Tang L, Huang J, et al. Roles of transforming growth factor-beta and phosphatidylinositol 3-kinase isoforms in integrin beta1-mediated bio-behaviors of mouse lung telocytes. J Transl Med 2019;17:431. [Crossref] [PubMed]
- Xiao P, Long X, Zhang L, et al. Neurotensin/IL-8 pathway orchestrates local inflammatory response and tumor invasion by inducing M2 polarization of Tumor-Associated macrophages and epithelial-mesenchymal transition of hepatocellular carcinoma cells. Oncoimmunology 2018;7:e1440166 [Crossref] [PubMed]
- Wang YY, Chen HD, Lo S, et al. Visfatin Enhances Breast Cancer Progression through CXCL1 Induction in Tumor-Associated Macrophages. Cancers (Basel) 2020;12:3526. [Crossref] [PubMed]
- Liguori M, Digifico E, Vacchini A, et al. The soluble glycoprotein NMB (GPNMB) produced by macrophages induces cancer stemness and metastasis via CD44 and IL-33. Cell Mol Immunol 2021;18:711-22. [Crossref] [PubMed]
- Lv J, Chen FK, Liu C, et al. Zoledronic acid inhibits thyroid cancer stemness and metastasis by repressing M2-like tumor-associated macrophages induced Wnt/beta-catenin pathway. Life Sci 2020;256:117925 [Crossref] [PubMed]
- Shao XJ, Xiang SF, Chen YQ, et al. Inhibition of M2-like macrophages by all-trans retinoic acid prevents cancer initiation and stemness in osteosarcoma cells. Acta Pharmacol Sin 2019;40:1343-50. [Crossref] [PubMed]
- Yang L, Dong Y, Li Y, et al. IL-10 derived from M2 macrophage promotes cancer stemness via JAK1/STAT1/NF-kappaB/Notch1 pathway in non-small cell lung cancer. Int J Cancer 2019;145:1099-110. [Crossref] [PubMed]
- Ning Y, Feng W, Cao X, et al. Genistein inhibits stemness of SKOV3 cells induced by macrophages co-cultured with ovarian cancer stem-like cells through IL-8/STAT3 axis. J Exp Clin Cancer Res 2019;38:19. [Crossref] [PubMed]
- Wei R, Zhu WW, Yu GY, et al. S100 calcium-binding protein A9 from tumor-associated macrophage enhances cancer stem cell-like properties of hepatocellular carcinoma. Int J Cancer 2021;148:1233-44. [Crossref] [PubMed]
- Jia J, Zhang H, Zhang H, et al. Infiltrating Macrophages Induced Stem-cell-like Features Through PI3K/AKT/GSK3beta Signaling to Promote Neurofibroma Growth. Arch Med Res 2020;51:124-34. [Crossref] [PubMed]
- Zhang X, Chen L, Dang WQ, et al. CCL8 secreted by tumor-associated macrophages promotes invasion and stemness of glioblastoma cells via ERK1/2 signaling. Lab Invest 2020;100:619-29. [Crossref] [PubMed]
- Chen Y, Wen H, Zhou C, et al. TNF-alpha derived from M2 tumor-associated macrophages promotes epithelial-mesenchymal transition and cancer stemness through the Wnt/beta-catenin pathway in SMMC-7721 hepatocellular carcinoma cells. Exp Cell Res 2019;378:41-50. [Crossref] [PubMed]
- Liu Z, Kuang W, Zhou Q, et al. TGF-β1 secreted by M2 phenotype macrophages enhances the stemness and migration of glioma cells via the SMAD2/3 signalling pathway. Int J Mol Med 2018;42:3395-403. [Crossref] [PubMed]
- Anfray C, Ummarino A, Andón FT, et al. Current Strategies to Target Tumor-Associated-Macrophages to Improve Anti-Tumor Immune Responses. Cells 2019;9:46. [Crossref] [PubMed]
- Jung KY, Cho SW, Kim YA, et al. Cancers with Higher Density of Tumor-Associated Macrophages Were Associated with Poor Survival Rates. J Pathol Transl Med 2015;49:318-24. [Crossref] [PubMed]
- Della Corte CM, Viscardi G, Papaccio F, et al. Implication of the Hedgehog pathway in hepatocellular carcinoma. World J Gastroenterol 2017;23:4330-40. [Crossref] [PubMed]
- Huynh J, Etemadi N, Hollande F, et al. The JAK/STAT3 axis: A comprehensive drug target for solid malignancies. Semin Cancer Biol 2017;45:13-22. [Crossref] [PubMed]
- Konishi J, Kawaguchi KS, Vo H, et al. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res 2007;67:8051-7. [Crossref] [PubMed]
- Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res 2006;66:605-12. [Crossref] [PubMed]
- Sica A, Allavena P, Mantovani A. Cancer related inflammation: the macrophage connection. Cancer Lett 2008;267:204-15. [Crossref] [PubMed]
- Zhang F, Wang H, Wang X, et al. TGF-beta induces M2-like macrophage polarization via SNAIL-mediated suppression of a pro-inflammatory phenotype. Oncotarget 2016;7:52294-306. [Crossref] [PubMed]
- Shen Z, Kauttu T, Cao J, et al. Macrophage coculture enhanced invasion of gastric cancer cells via TGF-beta and BMP pathways. Scand J Gastroenterol 2013;48:466-72. [Crossref] [PubMed]
- Xian G, Zhao J, Qin C, et al. Simvastatin attenuates macrophage-mediated gemcitabine resistance of pancreatic ductal adenocarcinoma by regulating the TGF-beta1/Gfi-1 axis. Cancer Lett 2017;385:65-74. [Crossref] [PubMed]
- Standiford TJ, Kuick R, Bhan U, et al. TGF-beta-induced IRAK-M expression in tumor-associated macrophages regulates lung tumor growth. Oncogene 2011;30:2475-84. [Crossref] [PubMed]
- Kawata M, Koinuma D, Ogami T, et al. TGF-beta-induced epithelial-mesenchymal transition of A549 lung adenocarcinoma cells is enhanced by pro-inflammatory cytokines derived from RAW 264.7 macrophage cells. J Biochem 2012;151:205-16. [Crossref] [PubMed]
- Yang XJ, Chen GL, Yu SC, et al. TGF-beta1 enhances tumor-induced angiogenesis via JNK pathway and macrophage infiltration in an improved zebrafish embryo/xenograft glioma model. Int Immunopharmacol 2013;15:191-8. [Crossref] [PubMed]
- Zhu C, Shen H, Zhu L, et al. Plasminogen Activator Inhibitor 1 Promotes Immunosuppression in Human Non-Small Cell Lung Cancers by Enhancing TGF-Beta1 Expression in Macrophage. Cell Physiol Biochem 2017;44:2201-11. [Crossref] [PubMed]
- Vainer N, Dehlendorff C, Johansen JS. Systematic literature review of IL-6 as a biomarker or treatment target in patients with gastric, bile duct, pancreatic and colorectal cancer. Oncotarget 2018;9:29820-41. [Crossref] [PubMed]
- Fu XL, Duan W, Su CY, et al. Interleukin 6 induces M2 macrophage differentiation by STAT3 activation that correlates with gastric cancer progression. Cancer Immunol Immunother 2017;66:1597-608. [Crossref] [PubMed]
- Yin Z, Ma T, Lin Y, et al. IL-6/STAT3 pathway intermediates M1/M2 macrophage polarization during the development of hepatocellular carcinoma. J Cell Biochem 2018;119:9419-32. [Crossref] [PubMed]
- Hsu HS, Lin JH, Hsu TW, et al. Mesenchymal stem cells enhance lung cancer initiation through activation of IL-6/JAK2/STAT3 pathway. Lung Cancer 2012;75:167-77. [Crossref] [PubMed]
- Zhang X, Hu F, Li G, et al. Human colorectal cancer-derived mesenchymal stem cells promote colorectal cancer progression through IL-6/JAK2/STAT3 signaling. Cell Death Dis 2018;9:25. [Crossref] [PubMed]
- Ibrahim SA, Gadalla R, El-Ghonaimy EA, et al. Syndecan-1 is a novel molecular marker for triple negative inflammatory breast cancer and modulates the cancer stem cell phenotype via the IL-6/STAT3, Notch and EGFR signaling pathways. Mol Cancer 2017;16:57. [Crossref] [PubMed]
- Xiong S, Wang R, Chen Q, et al. Cancer-associated fibroblasts promote stem cell-like properties of hepatocellular carcinoma cells through IL-6/STAT3/Notch signaling. Am J Cancer Res 2018;8:302-16. [PubMed]
- Qiu Y, Pu T, Guo P, et al. ALDH(+)/CD44(+) cells in breast cancer are associated with worse prognosis and poor clinical outcome. Exp Mol Pathol 2016;100:145-50. [Crossref] [PubMed]
- Ji W, Yu Y, Li Z, et al. FGFR1 promotes the stem cell-like phenotype of FGFR1-amplified non-small cell lung cancer cells through the Hedgehog pathway. Oncotarget 2016;7:15118-34. [Crossref] [PubMed]


